2N4E
 
 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
2MTL
 
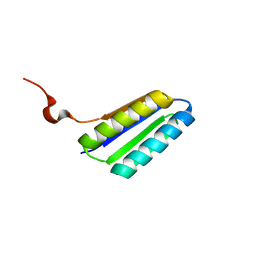 | | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109 | | Descriptor: | De novo designed protein FR55 OR109 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Ciccosanti, C, Sahdev, S, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed FR55, Northeast Structural Genomics Consortium (NESG) Target OR109
To be Published
|
|
2KPO
 
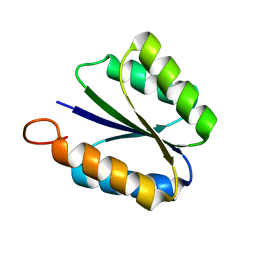 | | Solution NMR structure of de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium target OR16 | | Descriptor: | rossmann 2x2 fold protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Hamilton, K, Ciccosanti, C, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-17 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of denovo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium target OR16
To be Published
|
|
2MR6
 
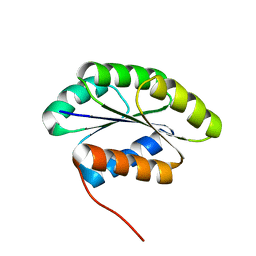 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR462 | | Descriptor: | De novo designed Protein OR462 | | Authors: | Xu, X, Nivon, L, Federizon, J.F, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR462
To be Published
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2N41
 
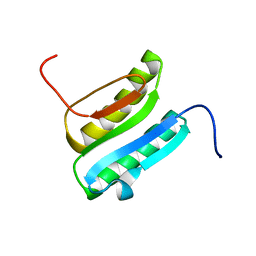 | | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34 | | Descriptor: | OR34 | | Authors: | Liu, G, Chan, K, Basanta, B, Xiao, R, Janjua, H, Kogan, S, Maglaqui, M, Ciccosanti, C, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of DE NOVO DESIGNED PROTEIN Top7NNSTYCC, Northeast Structural Genomics Consortium (NESG) Target OR34
To be Published
|
|
2N75
 
 | | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Rossmann2x2 Fold, Northeast Structural Genomics Consortium (NESG) Target OR446
To be Published
|
|
2L82
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR32 | | Descriptor: | DESIGNED PROTEIN OR32 | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Janjua, H, Tong, S, Acton, T.B, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target OR32
To be Published
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
9CKV
 
 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | De Novo Design of Integrin alpha 5 beta 1 Modulating Proteins to Enhance Biomaterial Properties.
Adv Mater, 2025
|
|
9DIA
 
 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin candidate 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-09-05 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | De Novo Design of Integrin alpha 5 beta 1 Modulating Proteins to Enhance Biomaterial Properties.
Adv Mater, 2025
|
|
9EF2
 
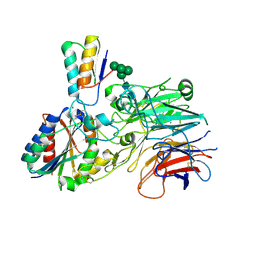 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin candidate 2, open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-11-19 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | De Novo Design of Integrin alpha 5 beta 1 Modulating Proteins to Enhance Biomaterial Properties.
Adv Mater, 2025
|
|
9BEI
 
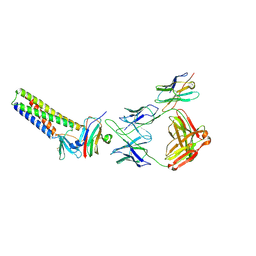 | |
7UHB
 
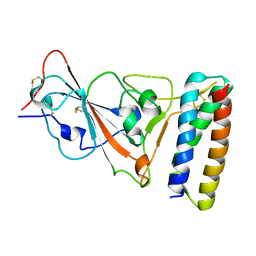 | |
7UHC
 
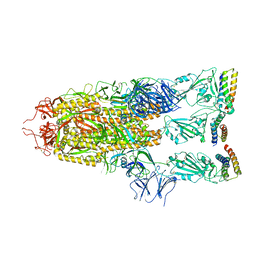 | |
7OPB
 
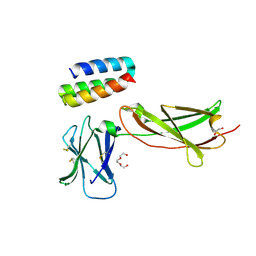 | | IL7R in complex with an antagonist | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IL7R binder, ... | | Authors: | Markovic, I, Verschueren, K.H.G, Verstraete, K, Savvides, S.N. | | Deposit date: | 2021-05-31 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
4W4M
 
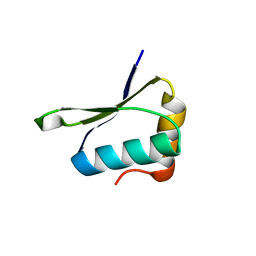 | | Crystal structure of PrgK 19-92 | | Descriptor: | Lipoprotein PrgK | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-08-15 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
3CQU
 
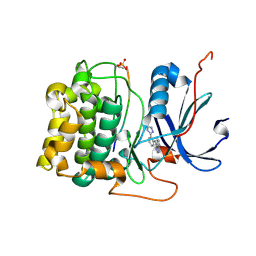 | | Crystal Structure of Akt-1 complexed with substrate peptide and inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[2-(5-methyl-4H-1,2,4-triazol-3-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-4-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Pandit, J. | | Deposit date: | 2008-04-03 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and structure based optimization of novel Akt inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7RDH
 
 | |
3J6D
 
 | | Model of the PrgH-PrgK periplasmic rings | | Descriptor: | Pathogenicity 1 island effector protein, Protein PrgH | | Authors: | Bergeron, J.R.C, Strynadka, N.C.J. | | Deposit date: | 2014-02-14 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | The Modular Structure of the Inner-Membrane Ring Component PrgK Facilitates Assembly of the Type III Secretion System Basal Body.
Structure, 23, 2015
|
|
8AH9
 
 | |
5JSB
 
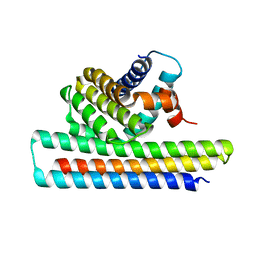 | | Crystal structure of Mcl1-inhibitor complex | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mcl-1 inhibitor | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2016-05-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Computationally designed high specificity inhibitors delineate the roles of BCL2 family proteins in cancer.
Elife, 5, 2016
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
7N1K
 
 | |
7N1J
 
 | |
