6B47
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF2 | | Descriptor: | Anti-CRISPR protein AcrF2, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6B48
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex with bound anti-CRISPR protein AcrF10 | | Descriptor: | Anti-CRISPR protein AcrF10, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
3IU3
 
 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
1XG7
 
 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
1XI8
 
 | | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001 | | Descriptor: | Molybdenum cofactor biosynthesis protein | | Authors: | Zhou, W, Zhao, M, Chang, J.C, Liu, Z.-J, Chen, L, Horanyi, P, Xu, H, Yang, H, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001
To be published
|
|
1XI6
 
 | | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001 | | Descriptor: | extragenic suppressor | | Authors: | Zhao, M, Chang, J.C, Zhou, W, Chen, L, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Extragenic suppressor from Pyrococcus furiosus Pfu-1862794-001
To be published
|
|
7W33
 
 | | The crystal structure of human CtsL in complex with 14a | | Descriptor: | N-[(2S)-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2R,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
7W34
 
 | | The crystal structure of human CtsL in complex with 14b | | Descriptor: | N-[(2S)-3-cyclohexyl-1-oxidanylidene-1-[[(2S,3S)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepiperidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-1-benzofuran-2-carboxamide, Procathepsin L | | Authors: | Zhao, Y, Shao, M, Zhao, J, Yang, H, Rao, Z. | | Deposit date: | 2021-11-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-based design of pan-coronavirus inhibitors targeting host cathepsin L and calpain-1.
Signal Transduct Target Ther, 9, 2024
|
|
5XQ1
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-3 | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8YFJ
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-ATTGGATCCAAT-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*AP*TP*TP*GP*GP*AP*TP*CP*CP*AP*AP*T)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YEY
 
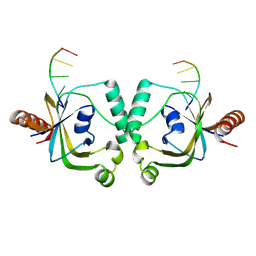 | | TRIP4 ASCH domain in complex with ssDNA-1 | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*TP*TP*TP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YFI
 
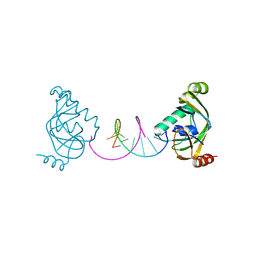 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCA-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YEW
 
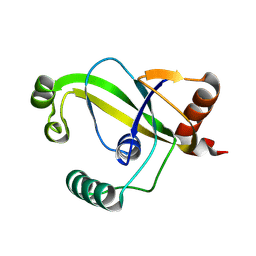 | | TRIP4 ASCH domain in unliganded form | | Descriptor: | Activating signal cointegrator 1 | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXX
 
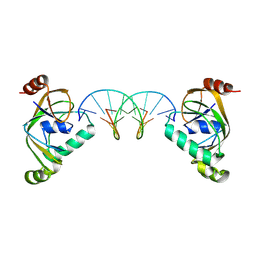 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCG-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*CP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*G)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXW
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCC-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
3IO8
 
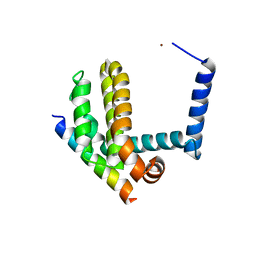 | | BimL12F in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
3GIZ
 
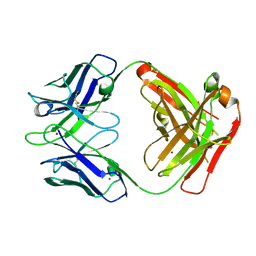 | | Crystal structure of the Fab fragment of anti-CD20 antibody Ofatumumab | | Descriptor: | Fab fragment of anti-CD20 antibody Ofatumumab, heavy chain, light chain, ... | | Authors: | Du, J, Yang, H, Ding, J. | | Deposit date: | 2009-03-07 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Fab fragment of therapeutic antibody Ofatumumab provides insights into the recognition mechanism with CD20
Mol.Immunol., 46, 2009
|
|
3IO9
 
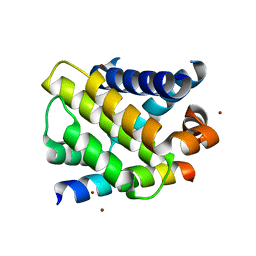 | | BimL12Y in complex with Mcl-1 | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Czabotar, P.E, Lee, E.F, Yang, H, Sleebs, B.E, Lessene, G, Colman, P.M, Smith, B.J, Fairlie, W.D. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
7VMU
 
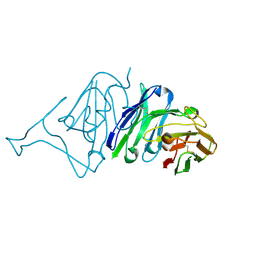 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | Spike protein S1, scFv E4 | | Authors: | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
7VBA
 
 | | Structure of the pre state human RNA Polymerase I Elongation Complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (5'-D(P*A*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*AP*GP*AP*CP*AP*GP*CP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBC
 
 | | Back track state of human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*G)-3'), DNA (5'-D(P*AP*GP*GP*AP*CP*AP*GP*CP*GP*TP*GP*TP*CP*AP*GP*CP*AP*AP*TP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7VBB
 
 | | Structure of the post state human RNA Polymerase I Elongation Complex | | Descriptor: | DNA (25-MER), DNA (5'-D(*CP*TP*GP*TP*CP*CP*TP*CP*TP*GP*GP*CP*GP*A)-3'), DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Zhao, D, Liu, W, Chen, K, Yang, H, Xu, Y. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structure of the human RNA polymerase I elongation complex.
Cell Discov, 7, 2021
|
|
7W7O
 
 | | The crystal structure of human Calpain-1 protease core in complex with 14a | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhao, J, Shao, M, Yang, H, Rao, Z. | | Deposit date: | 2021-12-06 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The crystal structure of human Calpain-1 protease core in complex with 14a
To Be Published
|
|
