7YMJ
 
 | | Cryo-EM structure of alpha1AAR-Nb6 complex bound to tamsulosin | | Descriptor: | Nb6, Tamsulosin, alpha1A-adrenergic receptor | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YMH
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to noradrenaline | | Descriptor: | Nb29, Noradrenaline, alpha1A-adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-28 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
7YM8
 
 | | Cryo-EM structure of Nb29-alpha1AAR-miniGsq complex bound to oxymetazoline | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Oxymetazoline, alpha1A adrenergic receptor, ... | | Authors: | Toyoda, Y, Zhu, A, Yan, C, Kobilka, B.K, Liu, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis of alpha 1A -adrenergic receptor activation and recognition by an extracellular nanobody.
Nat Commun, 14, 2023
|
|
6CAD
 
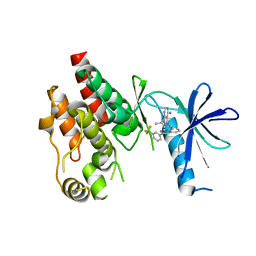 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
7XYD
 
 | | Crystal structure of TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Duan, Y, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-06-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
6C1H
 
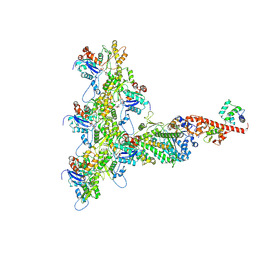 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1G
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3NR2
 
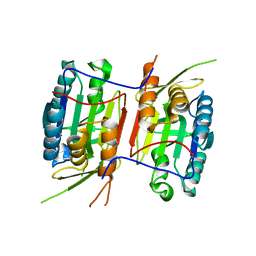 | | Crystal structure of Caspase-6 zymogen | | Descriptor: | Caspase-6 | | Authors: | Su, X.-D, Wang, X.-J, Liu, X, Mi, W, Wang, K.-T. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
3PMR
 
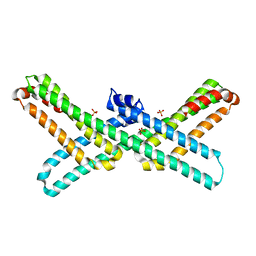 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
3V69
 
 | | Filia-N crystal structure | | Descriptor: | Protein Filia | | Authors: | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
3Q0E
 
 | | Crystals Structure of Aspartate beta-Semialdehyde Dehydrogenase from Vibrio Cholerae with product of S-allyl-L-cysteine sulfoxide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Liu, X, Viola, R.E. | | Deposit date: | 2010-12-15 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of inhibitors with selectivity against members of a homologous enzyme family.
Chem.Biol.Drug Des., 79, 2012
|
|
3URG
 
 | | The crystal structure of Anabaena CcbP | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, Alr1010 protein, SODIUM ION | | Authors: | Fan, X.X, Liu, X, Su, X.D. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Ellman's reagent in promoting crystallization and structure determination of Anabaena CcbP.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
9EQM
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-21 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[trans-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
9EQJ
 
 | | Crystal structure of pVHL:EloB:EloC in complex with MP-1-39 | | Descriptor: | (2S,4R)-1-[(2R)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-3-[[cis-4-(morpholin-4-ylmethyl)cyclohexyl]methylsulfanyl]butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kroupova, A, Pierri, M, Liu, X, Ciulli, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Stereochemical inversion at a 1,4-cyclohexyl PROTAC linker fine-tunes conformation and binding affinity.
Bioorg.Med.Chem.Lett., 110, 2024
|
|
3R5T
 
 | | Crystal structure of holo-ViuP | | Descriptor: | (4S,5R)-N-{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-N-[3-({[(4S,5R)-2-(2,3-dihydroxyphenyl)-5-met hyl-4,5-dihydro-1,3-oxazol-4-yl]carbonyl}amino)propyl]-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3OD5
 
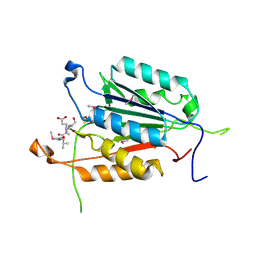 | | Crystal structure of active caspase-6 bound with Ac-VEID-CHO | | Descriptor: | CACODYLATE ION, Caspase-6, peptide aldehyde inhibitor AC-VEID-CHO | | Authors: | Wang, X.-J, Liu, X, Wang, K.-T, Cao, Q, Su, X.-D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation
Embo Rep., 11, 2010
|
|
3PYL
 
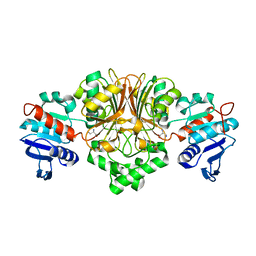 | | Crystal structure of aspartate beta-semialdehide dehydrogenase from Streptococcus pneumoniae with D-2,3-diaminopropionate | | Descriptor: | 3-amino-D-alanine, Aspartate-semialdehyde dehydrogenase | | Authors: | Pavlovsky, A.G, Liu, X, Faehnle, C.R, Potente, N, Viola, R.E. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
3R5S
 
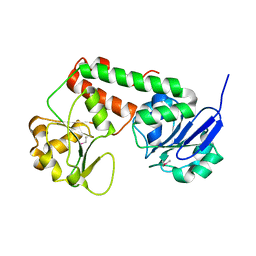 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
3WB8
 
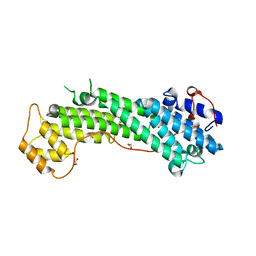 | | Crystal Structure of MyoVa-GTD | | Descriptor: | 1,2-ETHANEDIOL, Unconventional myosin-Va | | Authors: | Wei, Z, Liu, X, Yu, C, Zhang, M. | | Deposit date: | 2013-05-13 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis of cargo recognitions for class V myosins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7TIC
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TKU
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TI8
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TID
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7TIB
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the open sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*AP*GP*AP*CP*AP*CP*TP*AP*CP*GP*AP*GP*TP*AP*CP*AP*TP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*AP*TP*GP*TP*AP*CP*TP*CP*GP*TP*AP*GP*TP*GP*TP*CP*T)-3'), ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-13 | | Release date: | 2022-02-16 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
7THV
 
 | | Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) in an autoinhibited conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Pajak, J, Stone, N, Hayes, J, Demo, G, Kelch, B.A. | | Deposit date: | 2022-01-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures reveal high-resolution mechanism of a DNA polymerase sliding clamp loader.
Elife, 11, 2022
|
|
