8G85
 
 | | vFP52.02 Fab in complex with BG505 DS-SOSIP Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9W
 
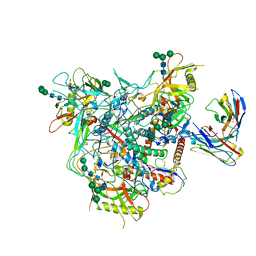 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9Y
 
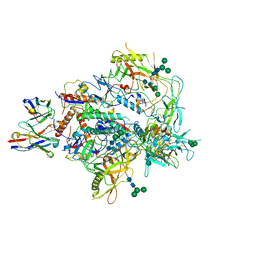 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
8G9X
 
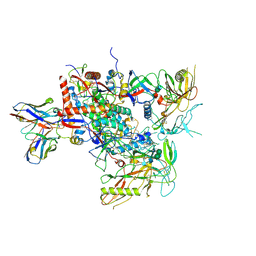 | | Cryo-EM structure of vFP49.02 Fab in complex with HIV-1 Env BG505 DS-SOSIP.664 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Changela, A, Gorman, J, Kwong, P.D. | | Deposit date: | 2023-02-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Diverse Murine Vaccinations Reveal Distinct Antibody Classes to Target Fusion Peptide and Variation in Peptide Length to Improve HIV Neutralization.
J.Virol., 97, 2023
|
|
4NRM
 
 | |
4NRO
 
 | |
4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | Descriptor: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | Authors: | Feng, C, Chen, Z, Liu, Y. | | Deposit date: | 2013-11-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
4NRP
 
 | |
4WK8
 
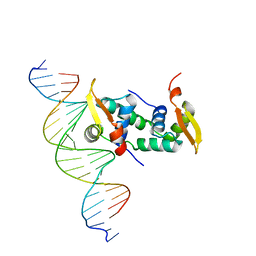 | | FOXP3 forms a domain-swapped dimer to bridge DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3'), Forkhead box protein P3 | | Authors: | Chen, Y, Chen, L. | | Deposit date: | 2014-10-01 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.
Nucleic Acids Res., 43, 2015
|
|
6NSL
 
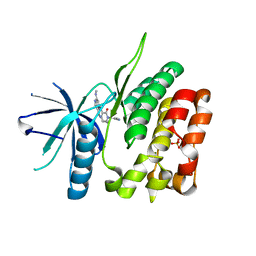 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH Compound-6c AKA 6-((1-(4-CYANOPHENY L)-2-OXO-1,2-DIHYDRO-3-PYRIDINYL)AMINO)-N-CYCLOPROPYL-8-(M ETHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBOXAMIDE | | Descriptor: | 6-{[1-(4-cyanophenyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}-N-cyclopropyl-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.M, Khan, J.A. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Imidazo[1,2-b]pyridazine Derivatives as Potent, Selective, and Orally Active Tyk2 JH2 Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | Descriptor: | GLYCEROL, Lon211 | | Authors: | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6E40
 
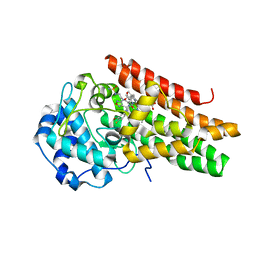 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complexed with ferric heme and Epacadostat | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E42
 
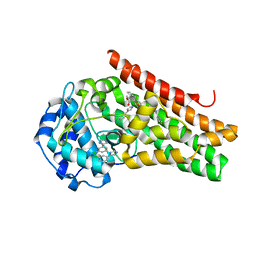 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and 4-Chlorophenyl imidazole | | Descriptor: | 4-(3-chlorophenyl)-1H-imidazole, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E41
 
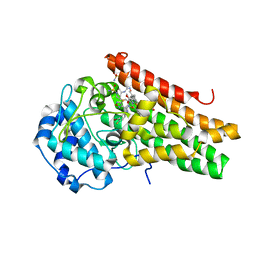 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and an Epacadostat analog | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]sulfanyl}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E43
 
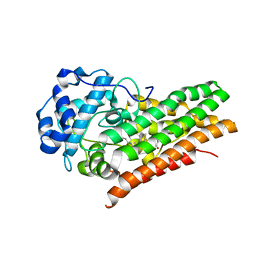 | | Crystal structure of human indoleamine 2,3-dioxygenase 1 (IDO1) in complex with a BMS-978587 analog | | Descriptor: | (1R,2S)-2-(4-[cyclohexyl(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, BENZOIC ACID, Indoleamine 2,3-dioxygenase 1 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E45
 
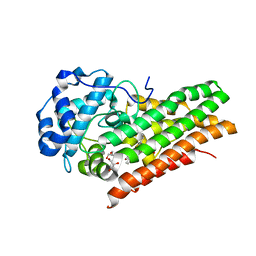 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferrous state | | Descriptor: | GLYCEROL, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E44
 
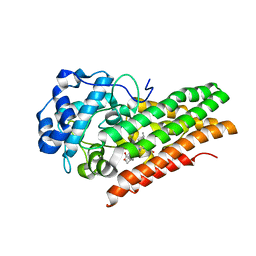 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferric state | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E46
 
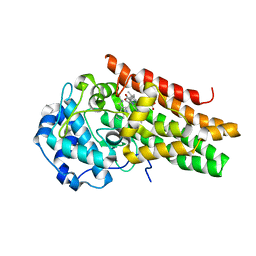 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferrous heme and tryptophan | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MVS
 
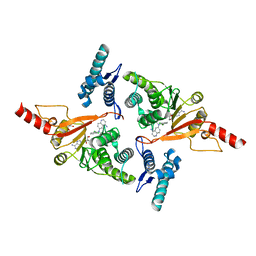 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW3
 
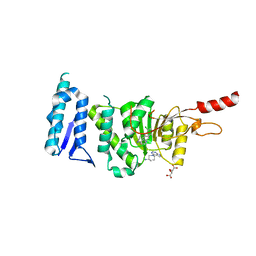 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
4NWK
 
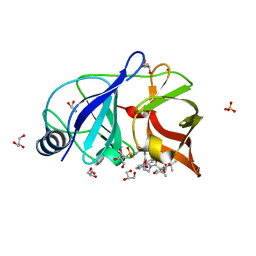 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-605339 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-n-((1r,2s)-1-((cyclopropylsulfonyl)carba moyl)-2-vinylcyclopropyl)-4-((6-methoxy-1-isoquinolinyl)ox y)-l-prolinamide | | Descriptor: | GLYCEROL, HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-4-[(6-methoxyisoquinolin-1-yl)oxy]-L-prolinamide, ... | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
4NT4
 
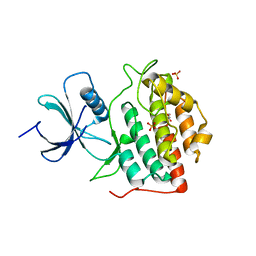 | | Crystal structure of the kinase domain of Gilgamesh isoform I from Drosophila melanogaster | | Descriptor: | GLYCEROL, Gilgamesh, isoform I, ... | | Authors: | Chen, C.C, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2013-11-30 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the kinase domain of Gilgamesh from Drosophila melanogaster
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8JYE
 
 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 and BTN2A1 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, Butyrophilin subfamily 2 member A1, ... | | Authors: | Yuan, L.J, Yang, Y.Y, Li, X, Cai, N.N, Chen, C.-C, Guo, R.-T, Zhang, Y.H. | | Deposit date: | 2023-07-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
