4FLP
 
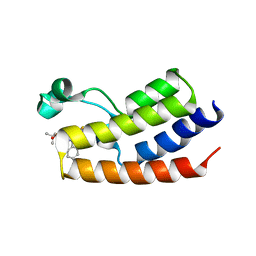 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain testis-specific protein, POTASSIUM ION | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Canning, P, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bradner, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-15 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Small-Molecule Inhibition of BRDT for Male Contraception.
Cell(Cambridge,Mass.), 150, 2012
|
|
4CGZ
 
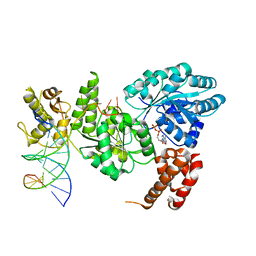 | | Crystal structure of the Bloom's syndrome helicase BLM in complex with DNA | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*CP*GP*AP*GP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*CP*TP*CP*GP*AP*CP*GP*CP*TP*CP*DT *CP*CP*CP)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Newman, J.A, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2013-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Bloom'S Syndrome Helicase Indicates a Role for the Hrdc Domain in Conformational Changes.
Nucleic Acids Res., 43, 2015
|
|
6TCX
 
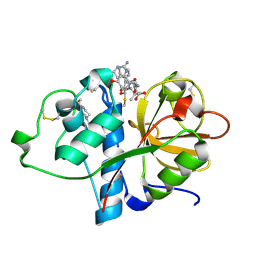 | | Papain bound to a natural cysteine protease inhibitor from Streptomyces mobaraensis | | Descriptor: | (2~{R})-2-[[(1~{S})-1-[(6~{S})-2-azanyl-1,4,5,6-tetrahydropyrimidin-6-yl]-2-[[(2~{S})-3-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]amino]-2-oxidanylidene-ethyl]carbamoylamino]-3-(4-hydroxyphenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Papain | | Authors: | Kraemer, A, Juettner, N.E, Fuchsbauer, H.-L, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding the Papain Inhibitor from Streptomyces mobaraensis as Being Hydroxylated Chymostatin Derivatives: Purification, Structure Analysis, and Putative Biosynthetic Pathway.
J.Nat.Prod., 83, 2020
|
|
4F7B
 
 | | Structure of the lysosomal domain of limp-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neculai, D, Ravichandran, M, Seitova, A, Neculai, M, Pizzaro, J.C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36.
Nature, 504, 2013
|
|
6TT5
 
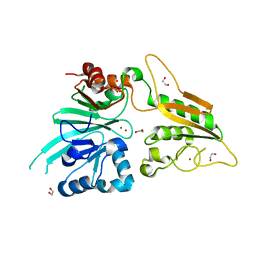 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
4DO0
 
 | | Crystal Structure of human PHF8 in complex with Daminozide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DAMINOZIDE, ... | | Authors: | Krojer, T, Daniel, M, Ng, S.S, Walport, L.J, Chowdhury, R, Arrowsmith, C.H, Edwards, A, Bountra, C, Kawamura, A, Muller-Knapp, S, McDonough, M.A, von Delft, F, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: |
|
|
4MEP
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3-chloro-pyridone ligand | | Descriptor: | 3-chloro-5-[1-(3-methylpyridin-2-yl)-3-phenyl-1H-1,2,4-triazol-5-yl]pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4MEO
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
6RRK
 
 | | Crystal structure of the central region of human cohesin subunit STAG1 in complex with RAD21 peptide | | Descriptor: | Cohesin subunit SA-1, Double-strand-break repair protein rad21 homolog | | Authors: | Newman, J.A, katis, V.L, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
Life Sci Alliance, 3, 2020
|
|
4CHB
 
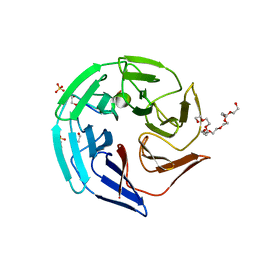 | | Crystal structure of the human KLHL2 Kelch domain in complex with a WNK4 peptide | | Descriptor: | 1,2-ETHANEDIOL, DODECAETHYLENE GLYCOL, KELCH-LIKE PROTEIN 2, ... | | Authors: | Sorrell, F.J, Schumacher, F.R, Kurz, T, Alessi, D.R, Newman, J, Cooper, C.D.O, Canning, P, Kopec, J, Williams, E, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-11-29 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Biochemical Characterisation of the Klhl3-Wnk Kinase Interaction Important in Blood Pressure Regulation.
Biochem.J., 460, 2014
|
|
6TSG
 
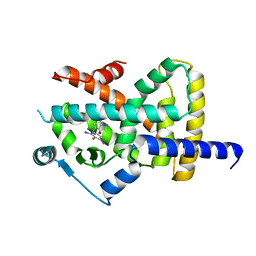 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG) in complex with TETRAC | | Descriptor: | 3,3',5,5'-TETRAIODOTHYROACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Gellrich, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | l-Thyroxin and the Nonclassical Thyroid Hormone TETRAC Are Potent Activators of PPAR gamma.
J.Med.Chem., 63, 2020
|
|
6SE4
 
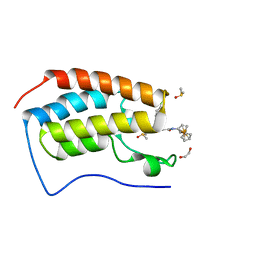 | | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor | | Descriptor: | (+)-JD1, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Hassell-Hart, S, Picaud, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Spencer, J, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with (+)-JD1, an Organometallic BET Bromodomain Inhibitor
To Be Published
|
|
6TU9
 
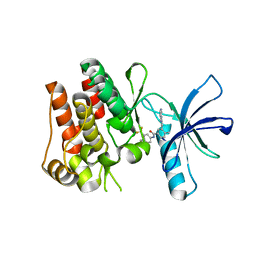 | | The ROR1 Pseudokinase Domain Bound To Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Inactive tyrosine-protein kinase transmembrane receptor ROR1 | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Niininen, W, Ungureanu, D, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
4FMU
 
 | | Crystal structure of Methyltransferase domain of human SET domain-containing protein 2 Compound: Pr-SNF | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-(propylamino)hexanoic acid, Histone-lysine N-methyltransferase SETD2, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zeng, H, Ibanez, G, Zheng, W, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
4FK7
 
 | | Crystal structure of Certhrax catalytic domain | | Descriptor: | CHLORIDE ION, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-12 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
6TUA
 
 | | The RYK Pseudokinase Domain | | Descriptor: | SULFATE ION, Tyrosine-protein kinase RYK | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Shin, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2020-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Insights into Pseudokinase Domains of Receptor Tyrosine Kinases.
Mol.Cell, 79, 2020
|
|
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
4CRH
 
 | | Crystal structure of the BTB-T1 domain of human SHKBP1 | | Descriptor: | SH3KBP1-BINDING PROTEIN 1 | | Authors: | Pinkas, D.M, Solcan, N, Krojer, T, Goubin, S, Williams, E.P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
4O62
 
 | | CW-type zinc finger of ZCWPW2 in complex with the amino terminus of histone H3 | | Descriptor: | Histone H3.3, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
4MI0
 
 | | Human Enhancer of Zeste (Drosophila) Homolog 2(EZH2) | | Descriptor: | Histone-lysine N-methyltransferase EZH2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Zeng, H, He, H, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations.
Plos One, 8, 2013
|
|
4QQN
 
 | | Protein arginine methyltransferase 3 in complex with compound MTV044246 | | Descriptor: | 1-{2-[1-(aminomethyl)cyclohexyl]ethyl}-3-isoquinolin-6-ylurea, CHLORIDE ION, GLYCEROL, ... | | Authors: | Dong, A, Dobrovetsky, E, Tempel, W, He, H, Zhao, K, Smil, D, Landon, M, Luo, X, Chen, Z, Dai, M, Yu, Z, Lin, Y, Zhang, H, Zhao, K, Schapira, M, Brown, P.J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Potent and Selective Allosteric Inhibitors of Protein Arginine Methyltransferase 3 (PRMT3).
J. Med. Chem., 61, 2018
|
|
4P7A
 
 | | Crystal Structure of human MLH1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Mlh1, MAGNESIUM ION, ... | | Authors: | Tempel, W, Lam, R, Zeng, H, Walker, J.R, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the human MLH1 N-terminus: implications for predisposition to Lynch syndrome.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4QSU
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with thymine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4QQG
 
 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
4QSS
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-methylpyrrolidin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
