2RA2
 
 | | X-Ray structure of the Q7CPV8 protein from Salmonella typhimurium at the resolution 1.9 A. Northeast Structural Genomics Consortium target StR88A | | Descriptor: | Putative lipoprotein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Vorobiev, S.M, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the Q7CPV8 protein from Salmonella typhimurium at the resolution 1.9 A.
To be Published
|
|
2RAD
 
 | | Crystal structure of the succinoglycan biosynthesis protein. Northeast structural Genomics Consortium target BcR135 | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the succinoglycan biosynthesis protein.
To be Published
|
|
5XDZ
 
 | | Crystal structure of zebrafish SNX25 PX domain | | Descriptor: | CHLORIDE ION, Cellular trafficking protein, SODIUM ION | | Authors: | Su, K, Zhang, Y, Xu, J, Liu, J. | | Deposit date: | 2017-03-30 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the PX domain of SNX25 reveals a novel phospholipid recognition model by dimerization in the PX domain
FEBS Lett., 591, 2017
|
|
3KLW
 
 | | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132. | | Descriptor: | Primosomal replication protein n | | Authors: | Kuzin, A.P, Neely, H, Vorobiev, S.M, Seetharaman, J, Forouhar, F, Wang, H, Janjua, H, Maglaqui, M, Xiao, T, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-09 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of primosomal replication protein n from Bordetella pertussis. northeast structural genomics consortium target ber132.
To be Published
|
|
3KW6
 
 | | Crystal Structure of a domain of 26S proteasome regulatory subunit 8 from homo sapiens. Northeast Structural Genomics Consortium target id HR3102A | | Descriptor: | 26S protease regulatory subunit 8 | | Authors: | Seetharaman, J, Su, M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a domain of 26S proteasome regulatory subunit 8 from homo sapiens. Northeast Structural Genomics Consortium target id HR3102A
To be Published
|
|
5WZS
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 8 | | Descriptor: | 2-[2-methyl-1-(naphthalen-1-ylmethyl)-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZM
 
 | | Crystal structure of human secreted phospholipase A2 group IIE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5WZV
 
 | | Crystal structure of human secreted phospholipase A2 group IIE with Me-indoxam | | Descriptor: | 2-[2-methyl-3-oxamoyl-1-[(2-phenylphenyl)methyl]indol-4-yl]oxyethanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
5XE1
 
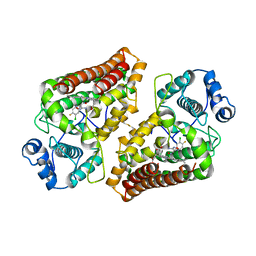 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with INCB14943 | | Descriptor: | 4-Amino-N-(3-chloro-4-fluorophenyl)-N'-hydroxy-1,2,5-oxadiazole-3-carboxamidine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, J, Wu, U, Liu, J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the binding mechanism of IDO1 with hydroxylamidine based inhibitor INCB14943
Biochem. Biophys. Res. Commun., 487, 2017
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2QGG
 
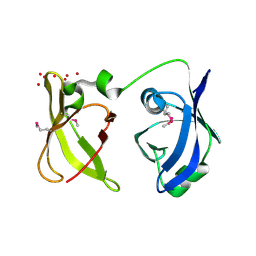 | | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248. Northeast Structural Genomics Consortium target AsR73. | | Descriptor: | 16S rRNA-processing protein rimM, POTASSIUM ION, UNKNOWN LIGAND | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-Ray structure of the protein Q6F7I0 from Acinetobacter calcoaceticus AmMS 248.
To be Published
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
4FDB
 
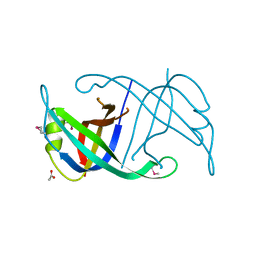 | | Three-dimensional structure of the protein prib from ralstonia solanacearum at the resolution 1.8a. northeast structural genomics consortium target rsr213c | | Descriptor: | ACETIC ACID, Probable primosomal replication protein n | | Authors: | Kuzin, A, Neely, H, Seetharaman, J, Wang, H, Sahdev, S, Foote, E.L, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of the protein prib from ralstonia solanacearum at the resolution 1.8a. northeast structural genomics consortium target rsr213c
To be Published
|
|
7YP6
 
 | | Crystal structure of elaiophylin glycosyltransferase in complex with UDP | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP3
 
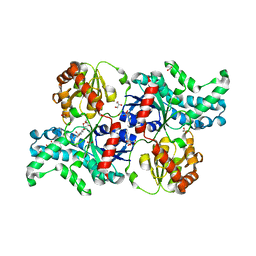 | | Crystal structure of elaiophylin glycosyltransferase in complex with elaiophylin | | Descriptor: | ACETATE ION, Elaiophylin, GLYCEROL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP5
 
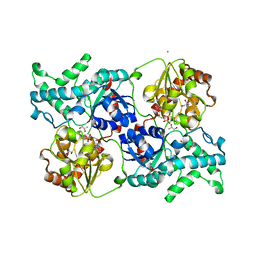 | | Crystal structure of elaiophylin glycosyltransferase in complex with TDP | | Descriptor: | CHLORIDE ION, Glycosyltransferase, R-1,2-PROPANEDIOL, ... | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7YP4
 
 | | Crystal structure of elaiophylin glycosyltransferase in apo-form | | Descriptor: | Glycosyltransferase, R-1,2-PROPANEDIOL | | Authors: | Xu, T, Liu, Q, Gan, Q, Liu, J. | | Deposit date: | 2022-08-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-induced dimerization of elaiophylin glycosyltransferase reveals a novel self-activating form of glycosyltransferase for symmetric glycosylation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4USG
 
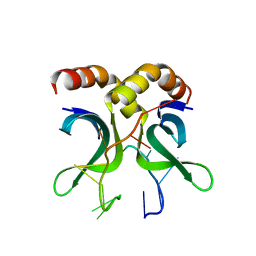 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
8HCU
 
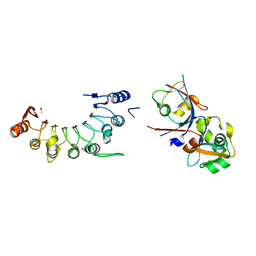 | | Crystal structure of BCOR/PCGF1/KDM2B complex | | Descriptor: | ACETIC ACID, BCL-6 corepressor, Polycomb group RING finger protein 1, ... | | Authors: | Shen, F, Chen, R, Xu, J, Liu, J. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of BCOR/PCGF1/KDM2B complex
to be published
|
|
8HQL
 
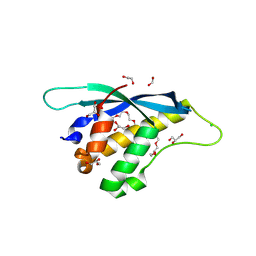 | |
3UAN
 
 | | Crystal structure of 3-O-sulfotransferase (3-OST-1) with bound PAP and heptasaccharide substrate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Moon, A.F, Xu, Y, Woody, S.M, Krahn, J.M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4XKL
 
 | | Crystal structure of NDP52 ZF2 in complex with mono-ubiquitin | | Descriptor: | ACETATE ION, Calcium-binding and coiled-coil domain-containing protein 2, GLYCEROL, ... | | Authors: | Xie, X, Li, F, Wang, Y, Lin, Z, Chen, X, Liu, J, Pan, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2
Autophagy, 11, 2015
|
|
4MWD
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) and ATP analog AMPPCP | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
4MVW
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1433) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
