8TLD
 
 | | Structure of the IL-5 Signaling Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, Interleukin-5, ... | | Authors: | Caveney, N.A, Garcia, K.C. | | Deposit date: | 2023-07-26 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Organizing structural principles of common beta family signaling
To Be Published
|
|
6W5Q
 
 | | Structure of the globular C-terminal domain of P. aeruginosa LpoP | | Descriptor: | Peptidoglycan synthase activator LpoP, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Caveney, N.A, Robb, C.S, Simorre, J.P, Strynadka, N.C.J. | | Deposit date: | 2020-03-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Peptidoglycan Synthase Activator LpoP in Pseudomonas aeruginosa.
Structure, 28, 2020
|
|
7LQ6
 
 | | CryoEM structure of Escherichia coli PBP1b | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2021-02-13 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
7KGN
 
 | | S. Typhi YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, L,D-transpeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
7KGM
 
 | | C. rodentium YcbB - ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Putative exported protein | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2020-10-17 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Cellular Insights into the l,d-Transpeptidase YcbB as a Therapeutic Target in Citrobacter rodentium, Salmonella Typhimurium, and Salmonella Typhi Infections.
Antimicrob.Agents Chemother., 65, 2021
|
|
7U7N
 
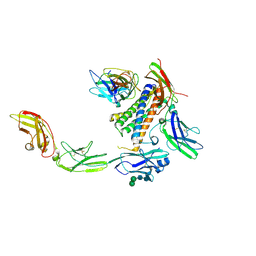 | | IL-27 quaternary receptor signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Caveney, N.A, Glassman, C.R, Jude, K.M, Tsutsumi, N, Garcia, K.C. | | Deposit date: | 2022-03-07 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the IL-27 quaternary receptor signaling complex.
Elife, 11, 2022
|
|
6NTZ
 
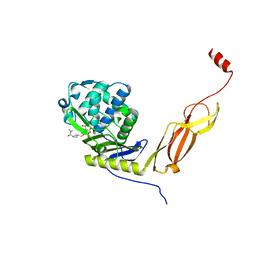 | | Crystal structure of E. coli PBP5-meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, D-alanyl-D-alanine carboxypeptidase | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
6N7O
 
 | | Crystal structure of GIL01 gp7 | | Descriptor: | GIL01 gp7, IODIDE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure, 27, 2019
|
|
6NTW
 
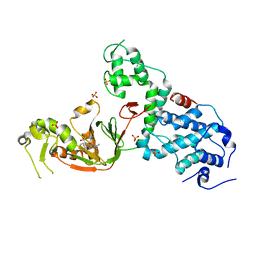 | | Crystal structure of E. coli YcbB | | Descriptor: | (2S,3R,4S)-4-{[(3S,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Probable L,D-transpeptidase YcbB, SULFATE ION | | Authors: | Caveney, N.A, Strynadka, N.C.J, Caballero, G, Worrall, L.J. | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-20 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural insight into YcbB-mediated beta-lactam resistance in Escherichia coli.
Nat Commun, 10, 2019
|
|
8EWY
 
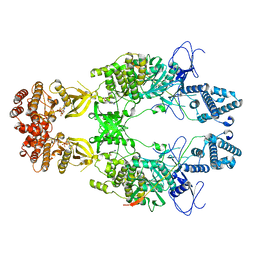 | | Structure of Janus Kinase (JAK) dimer complexed with cytokine receptor intracellular domain | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, Interferon lambda receptor 1, ... | | Authors: | Caveney, N.A, Saxton, R.A, Waghray, D, Garcia, K.C. | | Deposit date: | 2022-10-24 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural basis of Janus kinase trans-activation.
Cell Rep, 42, 2023
|
|
8FX4
 
 | | GC-C-Hsp90-Cdc37 regulatory complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Guanylyl cyclase C, Heat shock protein HSP 90-beta, ... | | Authors: | Caveney, N.A, Garcia, K.C. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into guanylyl cyclase receptor hijacking of the kinase-Hsp90 regulatory mechanism.
Elife, 12, 2023
|
|
6ZTG
 
 | | Spor protein DedD | | Descriptor: | Cell division protein DedD | | Authors: | Pazos, M, Peters, K, Boes, A, Safaei, Y, Kenward, C, Caveney, N.A, Laguri, C, Breukink, E, Strynadka, N.C.J, Simorre, J.P, Terrak, M, Vollmer, W. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | SPOR Proteins Are Required for Functionality of Class A Penicillin-Binding Proteins in Escherichia coli.
Mbio, 11, 2020
|
|
8VSE
 
 | | Cryo-EM structure of human CD45 extracellular region in complex with adenoviral protein E3/49K | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 45.5kDa protein, ... | | Authors: | Borowska, M.T, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-23 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Orientation-dependent CD45 inhibition with viral and engineered ligands.
Sci Immunol, 9, 2024
|
|
8VBW
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBU
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
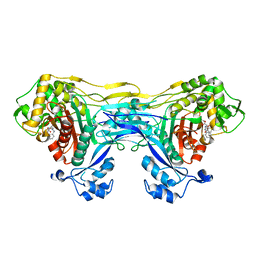 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VSP
 
 | | Cryo-EM structure of human invariant chain in complex with HLA-DQ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VRW
 
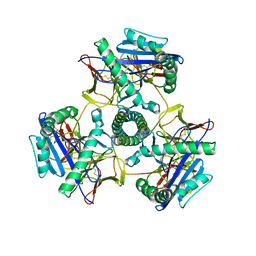 | | Cryo-EM structure of human invariant chain in complex with HLA-DR15 | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Wang, N, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2024-01-22 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into human MHC-II association with invariant chain.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VBT
 
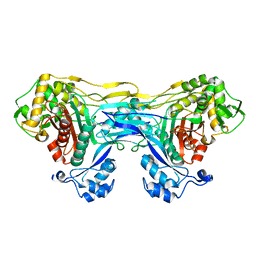 | |
8DHA
 
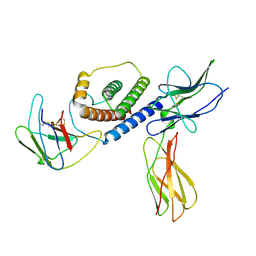 | |
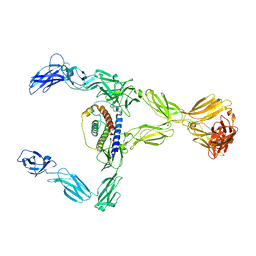
8DH8
 
 | |
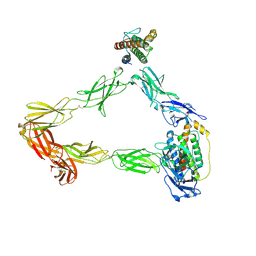
8DH9
 
 | |
7UWL
 
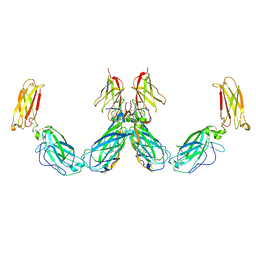 | | Structure of the IL-25-IL-17RB-IL-17RA ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWM
 
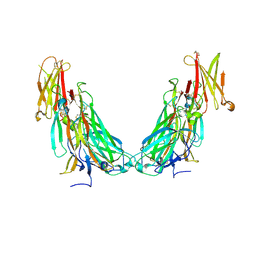 | | Structure of the IL-17A-IL-17RA binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor A, ... | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
7UWJ
 
 | | Structure of the homodimeric IL-25-IL-17RB binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-17 receptor B, Interleukin-25 | | Authors: | Wilson, S.C, Caveney, N.A, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Organizing structural principles of the IL-17 ligand-receptor axis.
Nature, 609, 2022
|
|
