1J3H
 
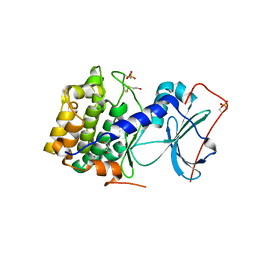 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
1L1F
 
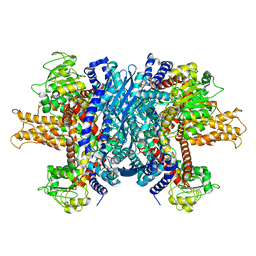 | | Structure of human glutamate dehydrogenase-apo form | | Descriptor: | Glutamate Dehydrogenase 1 | | Authors: | Smith, T.J, Schmidt, T, Fang, J, Wu, J, Siuzdak, G, Stanley, C.A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of apo human glutamate dehydrogenase details subunit communication and allostery.
J.Mol.Biol., 318, 2002
|
|
6VGU
 
 | |
1KKX
 
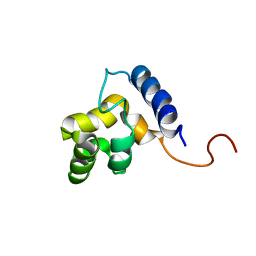 | | Solution structure of the DNA-binding domain of ADR6 | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-10 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1KN5
 
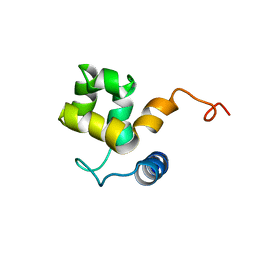 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1ME8
 
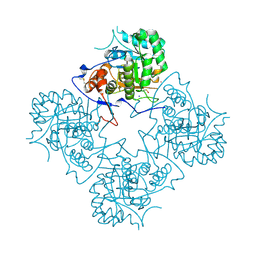 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, POTASSIUM ION, RIBAVIRIN MONOPHOSPHATE, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a
Catalysis-dependent Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1ME7
 
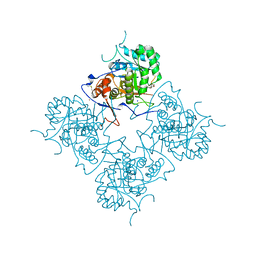 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP and MOA bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, MYCOPHENOLIC ACID, POTASSIUM ION, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a Catalysis-dependent
Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1PSU
 
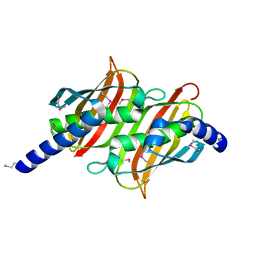 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | Descriptor: | Phenylacetic acid degradation protein PaaI | | Authors: | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-21 | | Release date: | 2003-07-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI.
J.Biol.Chem., 281, 2006
|
|
3QAL
 
 | | Crystal Structure of Arg280Ala mutant of Catalytic subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
3QAM
 
 | | Crystal Structure of Glu208Ala mutant of catalytic subunit of cAMP-dependent protein kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein kinase inhibitor, ... | | Authors: | Yang, J, Wu, J, Steichen, J, Taylor, S.S. | | Deposit date: | 2011-01-11 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
J.Mol.Biol., 415, 2012
|
|
6AGB
 
 | | Cryo-EM structure of yeast Ribonuclease P | | Descriptor: | RNases MRP/P 32.9 kDa subunit, Ribonuclease P RNA, Ribonuclease P protein subunit RPR2, ... | | Authors: | Lan, P, Tan, M, Wu, J, Lei, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insight into precursor tRNA processing by yeast ribonuclease P.
Science, 362, 2018
|
|
4ZOU
 
 | |
6BYM
 
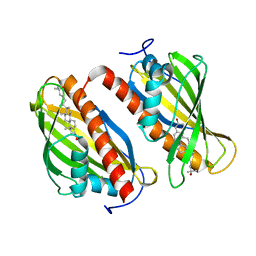 | | Crystal structure of the sterol-bound second StART domain of yeast Lam4 | | Descriptor: | 25-HYDROXYCHOLESTEROL, Sterol-binding protein | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
4OV9
 
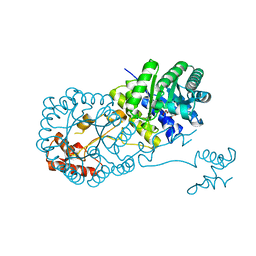 | | Structure of isopropylmalate synthase binding with alpha-isopropylmalate | | Descriptor: | (2S)-2-hydroxy-2-(propan-2-yl)butanedioic acid, ZINC ION, isopropylmalate synthase | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
4OV4
 
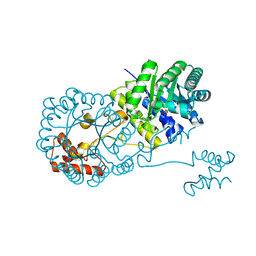 | | Isopropylmalate synthase binding with ketoisovalerate | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, ZINC ION | | Authors: | Zhang, Z, Wu, J, Wang, C, Zhang, P. | | Deposit date: | 2014-02-20 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subdomain II of alpha-isopropylmalate synthase is essential for activity: inferring a mechanism of feedback inhibition.
J.Biol.Chem., 289, 2014
|
|
6BYD
 
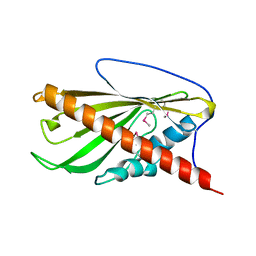 | | Crystal structure of the second StART domain of yeast Lam4 | | Descriptor: | Membrane-anchored lipid-binding protein LAM4 | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-31 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
4G84
 
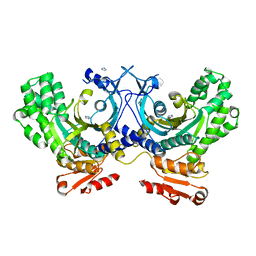 | | Crystal structure of human HisRS | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic, ... | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
5XXE
 
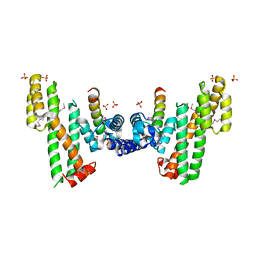 | | Crystal structure of Poz1 and Tpz1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, SULFATE ION, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
4G85
 
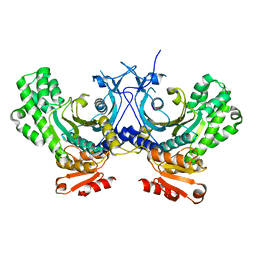 | | Crystal structure of human HisRS | | Descriptor: | Histidine-tRNA ligase, cytoplasmic | | Authors: | Wei, Z, Wu, J, Zhou, J.J, Yang, X.-L, Zhang, M, Schimmel, P. | | Deposit date: | 2012-07-21 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Internally Deleted Human tRNA Synthetase Suggests Evolutionary Pressure for Repurposing.
Structure, 20, 2012
|
|
5XXF
 
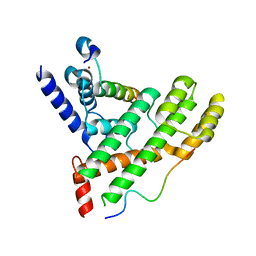 | | Crystal structure of Poz1, Tpz1 and Rap1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, Rap1, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
2JXW
 
 | |
2M80
 
 | | Solution structure of yeast dithiol glutaredoxin Grx8 | | Descriptor: | Glutaredoxin-8 | | Authors: | Tang, Y, Zhang, J, Yu, J, Wu, J, Zhou, C.Z, Shi, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-guided activity enhancement and catalytic mechanism of yeast grx8
Biochemistry, 53, 2014
|
|
4GMV
 
 | |
4GN1
 
 | | Crystal Structure of the RA and PH domains of Lamellipodin | | Descriptor: | MALONATE ION, Ras-associated and pleckstrin homology domains-containing protein 1 | | Authors: | Chang, Y.C.E, Wu, J. | | Deposit date: | 2012-08-16 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Lamellipodin implicates diverse functions in actin polymerization and Ras signaling.
Protein Cell, 4, 2013
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
