8XCG
 
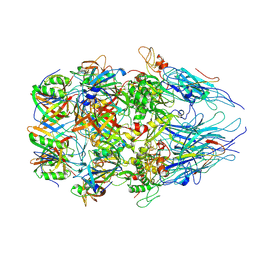 | | Tail tip complex of bacteriophage lambda in the open state | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Ge, X.F, Wang, J.W. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mechanism of bacteriophage lambda tail's interaction with the bacterial receptor.
Nat Commun, 15, 2024
|
|
8XCI
 
 | |
8XCK
 
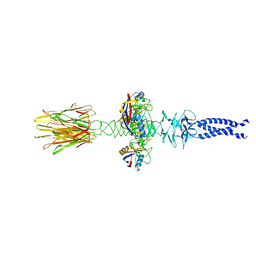 | | Closed state of central tail fiber of bacteriophage lambda | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, Tip attachment protein J | | Authors: | Ge, X.F, Wang, J.W. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural mechanism of bacteriophage lambda tail's interaction with the bacterial receptor.
Nat Commun, 15, 2024
|
|
8XCJ
 
 | |
6AX1
 
 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-yl 3-(3-phenyl-1,2,4-oxadiazol-5-yl)azetidine-1-carboxylate, GLYCEROL, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Azetidine and Piperidine Carbamates as Efficient, Covalent Inhibitors of Monoacylglycerol Lipase.
J. Med. Chem., 60, 2017
|
|
7YEU
 
 | | Superfolder green fluorescent protein with phosphine unnatural amino acid P3BF | | Descriptor: | Superfolder green fluorescent protein | | Authors: | Hu, C, Duan, H.Z, Liu, X.H, Chen, Y.X, Wang, J.Y. | | Deposit date: | 2022-07-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetically Encoded Phosphine Ligand for Metalloprotein Design.
J.Am.Chem.Soc., 144, 2022
|
|
3WMB
 
 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 | | Descriptor: | 2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
4HCN
 
 | | Crystal structure of Burkholderia pseudomallei effector protein CHBP in complex with ubiquitin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Yao, Q, Cui, J, Zhu, Y, Shao, F. | | Deposit date: | 2012-09-30 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ubiquitin and NEDD8 deamidation catalyzed by bacterial effectors that induce macrophage-specific apoptosis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HCP
 
 | |
7K4I
 
 | | Human Arginase 1 in complex with compound 06. | | Descriptor: | 3-[(1~{S},2~{S},5~{R})-2-carboxy-6-thia-3-azabicyclo[3.2.0]heptan-1-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4H
 
 | | Human Arginase 1 in complex with compound 04. | | Descriptor: | 3-[(1~{R},5~{S},8~{R})-5-carboxy-2,6-diazabicyclo[3.2.1]octan-8-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4J
 
 | | Human Arginase 1 in complex with compound 51. | | Descriptor: | 3-[(1~{S},2~{S},5~{R})-2-carboxy-3,6-diazabicyclo[3.2.0]heptan-1-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4G
 
 | | Human Arginase 1 in complex with compound 01. | | Descriptor: | 3-[(2~{S},3~{R})-2-carboxypyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4K
 
 | | Human Arginase 1 in complex with compound 52. | | Descriptor: | 3-[(3~{a}~{S},4~{S},6~{a}~{R})-4-carboxy-2,3,4,5,6,6~{a}-hexahydro-1~{H}-pyrrolo[2,3-c]pyrrol-3~{a}-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5C8T
 
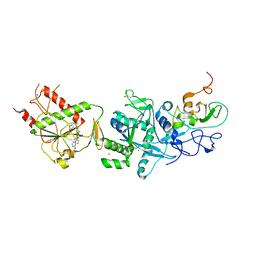 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex with functional ligand SAM | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1DAR
 
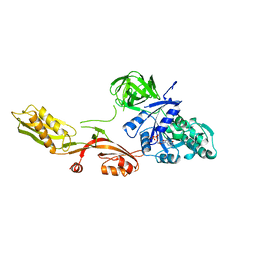 | | ELONGATION FACTOR G IN COMPLEX WITH GDP | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Al-Karadaghi, S, Aevarsson, A, Garber, M, Zheltonosova, J, Liljas, A. | | Deposit date: | 1996-02-15 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of elongation factor G in complex with GDP: conformational flexibility and nucleotide exchange.
Structure, 4, 1996
|
|
8GWO
 
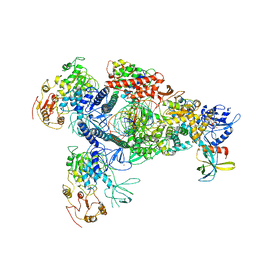 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
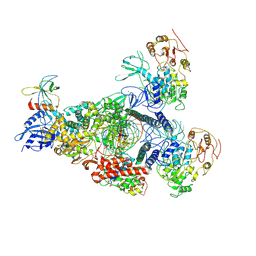 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
5C8U
 
 | | Crystal structure of the SARS coronavirus nsp14-nsp10 complex | | Descriptor: | Guanine-N7 methyltransferase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Ma, Y.Y, Wu, L.J, Zhang, R.G, Rao, Z.H. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3ZTX
 
 | | Aurora kinase selective inhibitors identified using a Taxol-induced checkpoint sensitivity screen. | | Descriptor: | 2-((4-(4-HYDROXYPIPERIDIN-1-YL)PHENYL)AMINO)-5,11-DIMETHYL-5H-BENZO[E]PYRIMIDO [5,4-B][1,4]DIAZEPIN-6(11H)-ONE, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Kwiatkowski, N, Villa, F, Musacchio, A, Gray, N. | | Deposit date: | 2011-07-12 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Aurora Kinase Inhibitors Identified Using a Taxol- Induced Checkpoint Sensitivity Screen.
Acs Chem.Biol., 7, 2012
|
|
8GTK
 
 | |
8GTJ
 
 | |
8GTN
 
 | |
