7F5A
 
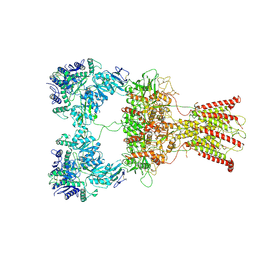 | | DNQX-bound GluK2-2xNeto2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
6K9N
 
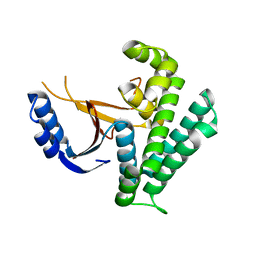 | | Rice_OTUB_like_catalytic domain | | Descriptor: | Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
5CQJ
 
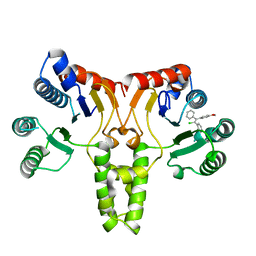 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase in complex with clomiphene | | Descriptor: | Clomifene, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQB
 
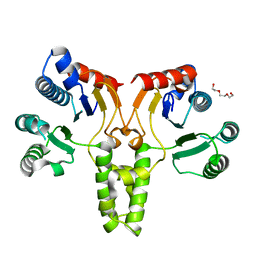 | | Crystal structure of E. coli undecaprenyl pyrophosphate synthase | | Descriptor: | Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), TRIETHYLENE GLYCOL | | Authors: | Worrall, L.J, Conrady, D.G, Strynadka, N.C. | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6N1W
 
 | |
4QO4
 
 | |
5UG9
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5UGA
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium | | Descriptor: | 4-(4-{[2-{[(3S)-1-acetylpyrrolidin-3-yl]amino}-9-(propan-2-yl)-9H-purin-6-yl]amino}phenyl)-1-methylpiperazin-1-ium, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
3KFA
 
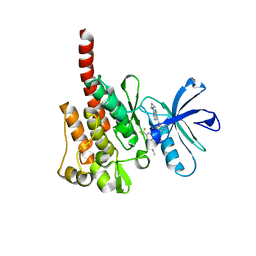 | |
5UGB
 
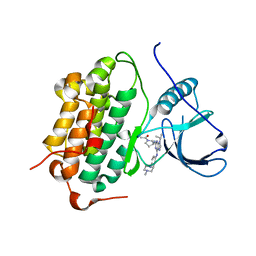 | |
5VAM
 
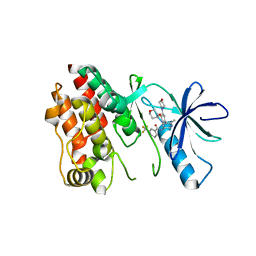 | | BRAF in Complex with RAF709 | | Descriptor: | N-{2-methyl-5'-(morpholin-4-yl)-6'-[(oxan-4-yl)oxy][3,3'-bipyridin]-5-yl}-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
7VKA
 
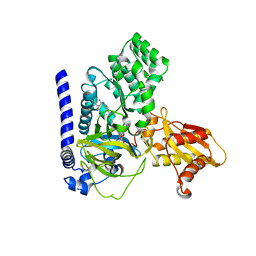 | | Crystal Structure of GH3.6 in complex with an inhibitor | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Indole-3-acetic acid-amido synthetase GH3.6, ... | | Authors: | Wang, N, Luo, M, Bao, H, Huang, H. | | Deposit date: | 2021-09-29 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5VAL
 
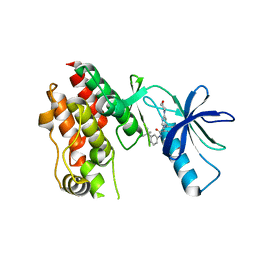 | | BRAF in Complex with N-(3-(tert-butyl)phenyl)-4-methyl-3-(6-morpholinopyrimidin-4-yl)benzamide | | Descriptor: | N-(3-tert-butylphenyl)-4-methyl-3-[6-(morpholin-4-yl)pyrimidin-4-yl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
3LLR
 
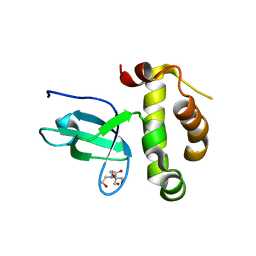 | | Crystal structure of the PWWP domain of Human DNA (cytosine-5-)-methyltransferase 3 alpha | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (cytosine-5)-methyltransferase 3A, SULFATE ION | | Authors: | Qiu, W, Dombrovski, L, Ni, S, Weigelt, J, Boutra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3LDN
 
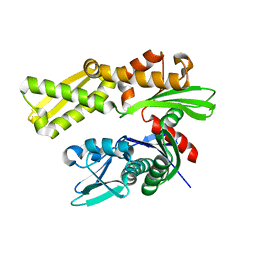 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in apo form | | Descriptor: | 78 kDa glucose-regulated protein | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3LDQ
 
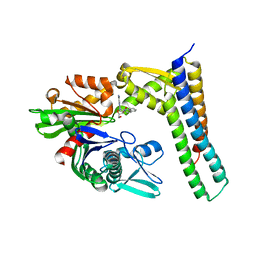 | | Crystal structure of HSC70/BAG1 in complex with small molecule inhibitor | | Descriptor: | 8-[(quinolin-2-ylmethyl)amino]adenosine, BAG family molecular chaperone regulator 1, Heat shock cognate 71 kDa protein | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
3LDP
 
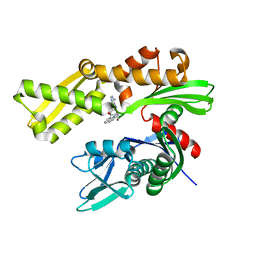 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with small molecule inhibitor | | Descriptor: | 78 kDa glucose-regulated protein, 8-[(quinolin-2-ylmethyl)amino]adenosine | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
7FCV
 
 | | Cryo-EM structure of the Potassium channel AKT1 mutant from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Jia, Y.T, Zhang, Y.M, Tang, R.F, Xu, X, Li, X.M, Lei, J.L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
6K8K
 
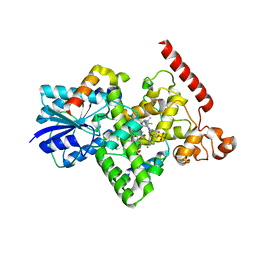 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8IFG
 
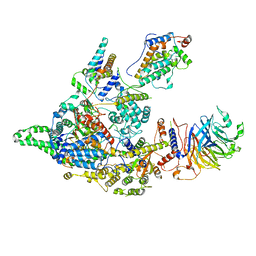 | | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Cph2, ... | | Authors: | Zhang, H.Q, Wang, X, Wang, Y.N, Liu, S.M, Zhang, Y, Xu, K, Ji, L.T, Kornberg, R.D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Class I histone deacetylase complex: Structure and functional correlates.
Proc Natl Acad Sci U S A, 120, 2023
|
|
6KK1
 
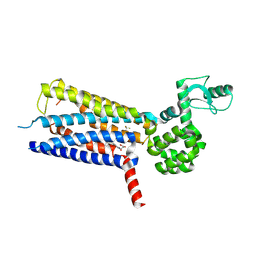 | | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
3LDL
 
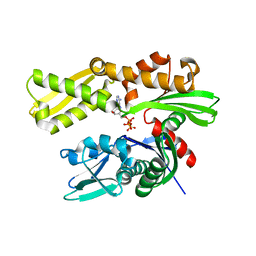 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Dokurno, P, Surgenor, A.E, Shaw, T, Macias, A.T, Massey, A.J, Williamson, D.S. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adenosine-Derived Inhibitors of 78 kDa Glucose Regulated Protein (Grp78) ATPase: Insights into Isoform Selectivity.
J.Med.Chem., 54, 2011
|
|
8J63
 
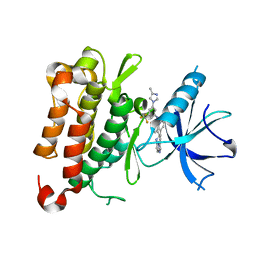 | | The crystal structure of TrkA kinase in complex with 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide | | Descriptor: | 4^6-methyl-N-(3-(4-methyl-1H-imidazol-1-yl)-5-(trifluoromethyl)phenyl)-11-oxo-5-oxa-10,14-diaza-1(3,6)-imidazo[1,2-b]pyridazina-4(1,3)-benzenacyclotetradecaphan-2-yne-4^5-carboxamide, High affinity nerve growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.0005 Å) | | Cite: | Structure-Based Optimization of the Third Generation Type II Macrocycle TRK Inhibitors with Improved Activity against Solvent-Front, xDFG, and Gatekeeper Mutations.
J.Med.Chem., 66, 2023
|
|
6K8I
 
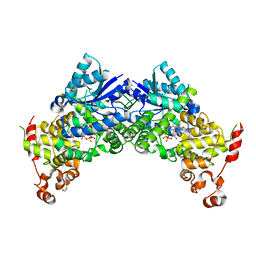 | | Crystal structure of Arabidopsis thaliana CRY2 | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, L, Wang, X, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
