5Y8Y
 
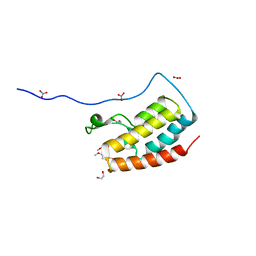 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
7YQE
 
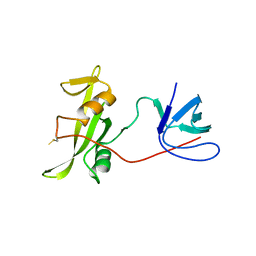 | |
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8W
 
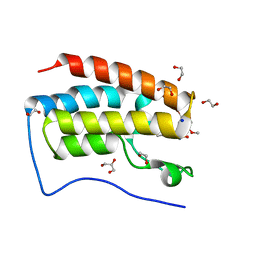 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-N-(3-methyl-6-oxidanyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y94
 
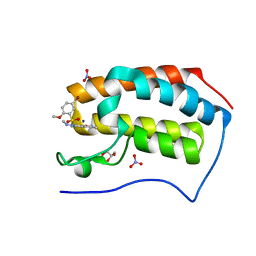 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 5-bromanyl-2-methoxy-N-[3-methyl-6-(methylamino)-1,2-benzoxazol-5-yl]benzenesulfonamide, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8Z
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-N-(3,6-dimethyl-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
5Y8C
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloranyl-2-methoxy-N-(6-methoxy-3-methyl-1,2-benzoxazol-5-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-21 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
6JCJ
 
 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9CSI
 
 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
8K1O
 
 | | mycobacterial efflux pump, AMPPNP bound state | | Descriptor: | CARDIOLIPIN, MAGNESIUM ION, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K1N
 
 | | mycobacterial efflux pump, substrate-bound state | | Descriptor: | CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, Multidrug efflux system permease protein Rv1217c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K1M
 
 | | mycobacterial efflux pump, apo state | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CARDIOLIPIN, Multidrug efflux system ATP-binding protein Rv1218c, ... | | Authors: | Wang, Y, Wu, F, Zhang, L, Rao, Z. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of a mycobacterial ABC transporter that mediates rifampicin resistance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5XXH
 
 | | Crystal Structure Analysis of the CBP | | Descriptor: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
7YAC
 
 | | Paltusotine-bound SSTR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, J, Shao, Z. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
7YAE
 
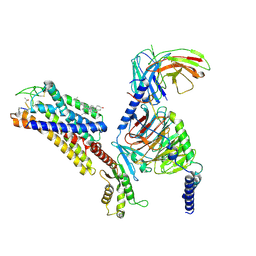 | | Octreotide-bound SSTR2-Gi complex | | Descriptor: | CHOLESTEROL, DPN-CYS-PHE-DTR-LYS-THR-CYS-THO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, J, Shao, Z. | | Deposit date: | 2022-06-28 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Prospect of acromegaly therapy: molecular mechanism of clinical drugs octreotide and paltusotine.
Nat Commun, 14, 2023
|
|
8C3A
 
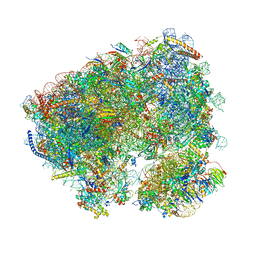 | | Crystal structure of ailanthone bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New crystal system to promote screening for new eukaryotic inhibitors
To Be Published
|
|
8SAI
 
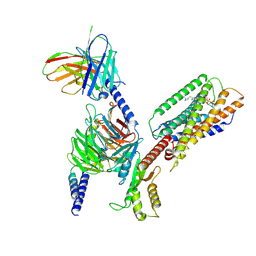 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9NSE
 
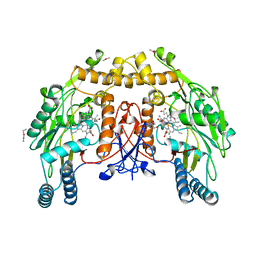 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, ETHYL-ISOSELENOUREA COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLIC ACID, ... | | Authors: | Li, H, Raman, C.S, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2000-10-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Mapping the active site polarity in structures of endothelial nitric oxide synthase heme domain complexed with isothioureas.
J.Inorg.Biochem., 81, 2000
|
|
1ED5
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH NNA(H4B FREE) | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-01-26 | | Release date: | 2001-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
1NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND SUBSTRATE L-ARGININE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
3NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND PRODUCT ANALOGUE L-THIOCITRULLINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, L-THIOCITRULLINE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-06 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
9ERY
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-[bis(fluoranyl)methyl]-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]methoxy]-2-[(2-hydroxyethylamino)methyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Plewka, J, Magiera-Mularz, K, Zhang, W. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of antitumor activity of 2-arylmethoxy-4-(2-fluoromethyl-biphenyl-3-ylmethoxy) benzylamine derivatives as PD-1/PD-l1 inhibitors.
Eur.J.Med.Chem., 276, 2024
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
5XLZ
 
 | |
