6BOZ
 
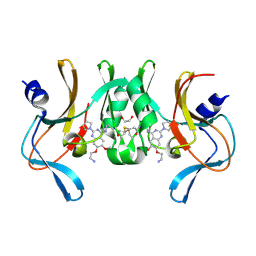 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
6JV5
 
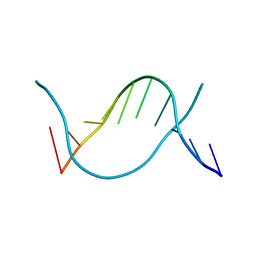 | | Crystal structure of 5-methylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(5CM)P*GP*CP*TP*GP*G)-3') | | Authors: | Zhang, L, Wang, Y.X. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5DEU
 
 | | Crystal structure of TET2-5hmC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DNA (5'-D(*AP*CP*CP*AP*CP*(5HC)P*GP*GP*TP*GP*GP*T)-3'), ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-26 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
5D9Y
 
 | | Crystal structure of TET2-5fC complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*(5FC)P*GP*AP*AP*GP*CP*T)-3'), DNA (5'-D(*AP*GP*CP*TP*TP*CP*GP*AP*CP*AP*GP*T)-3'), FE (III) ION, ... | | Authors: | Hu, L, Cheng, J, Rao, Q, Li, Z, Li, J, Xu, Y. | | Deposit date: | 2015-08-19 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Structural insight into substrate preference for TET-mediated oxidation.
Nature, 527, 2015
|
|
8E86
 
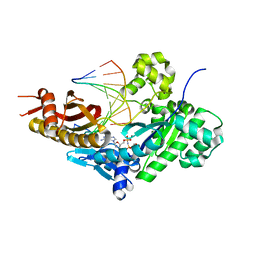 | | Human DNA polymerase eta-DNA-rC-ended primer-dGMPNPP ternary mismatch complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8B
 
 | |
8E8A
 
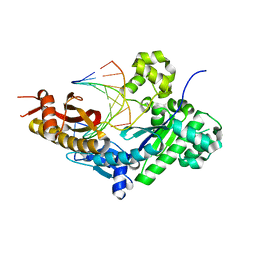 | | Human DNA polymerase eta-DNA-dT-ended primer binary complex | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E87
 
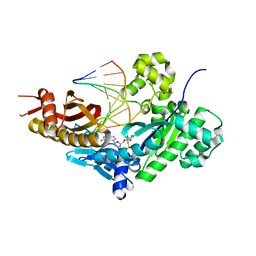 | | Human DNA polymerase eta-DNA-rA-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*TP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8J
 
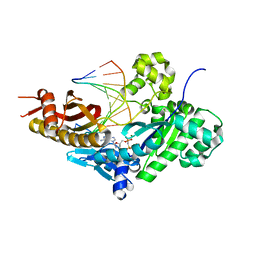 | | Human DNA polymerase eta-DNA-rG-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*CP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8G
 
 | |
8E8D
 
 | |
8E85
 
 | | Human DNA polymerase eta-DNA-rG-ended primer-dGMPNPP ternary mismatch complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*CP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8F
 
 | |
8E8C
 
 | |
8E89
 
 | | Human DNA polymerase eta-DNA-rU-ended primer-binary complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, DNA/RNA (5'-D(*AP*GP*CP*GP*TP*CP*A)-R(P*U)-3') | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8H
 
 | |
8E88
 
 | | Human DNA polymerase eta-DNA-rU-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8K
 
 | | Human DNA polymerase eta-DNA-rC-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8E
 
 | |
7MON
 
 | | Structure of human RIPK3-MLKL complex | | Descriptor: | Mixed lineage kinase domain-like protein, N-[4-({2-[(cyclopropanecarbonyl)amino]pyridin-4-yl}oxy)-3-fluorophenyl]-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Meng, Y, Davies, K.A, Czabotar, P.E, Murphy, J.M. | | Deposit date: | 2021-05-03 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
7MX3
 
 | | Crystal structure of human RIPK3 complexed with GSK'843 | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-5-yl)-7-(1,3-dimethyl-1H-pyrazol-5-yl)thieno[3,2-c]pyridin-4-amine, Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Davies, K.A, Czabotar, P.E. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis.
Nat Commun, 12, 2021
|
|
8SPE
 
 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P31 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SPZ
 
 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with dioctanoyl phosphatidylserine | | Descriptor: | Apoptosis regulator BAX, SULFATE ION | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E, Miller, M.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SVK
 
 | | Crystal structure of Bax D71N core domain BH3-groove dimer | | Descriptor: | Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Miller, M.S, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2023-05-16 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
8SPF
 
 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with 2-stearoyl lysoPC | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DODECANE, ... | | Authors: | Cowan, A.D, Miller, M.S, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
