5JHL
 
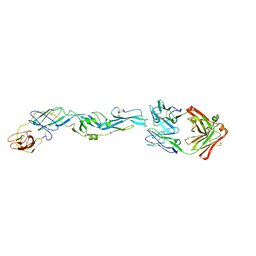 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | Descriptor: | Antibody Heavy chain, antibody Light chain, envelope protein | | Authors: | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
3OY7
 
 | |
6JJB
 
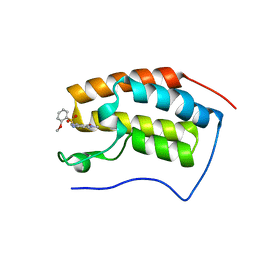 | | BRD4 in complex with ZZM1 | | Descriptor: | 2-methoxy-N-(1-methyl-2-oxidanylidene-benzo[cd]indol-6-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
3OY2
 
 | |
4EJF
 
 | |
3SWJ
 
 | | Crystal structure of Campylobacter jejuni ChuZ | | Descriptor: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | Authors: | Hu, Y. | | Deposit date: | 2011-07-14 | | Release date: | 2011-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
3NB6
 
 | |
5ZZ3
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | Descriptor: | Butyrophilin, subfamily 3, member A3 isoform b variant | | Authors: | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
4K6G
 
 | | Crystal structure of CALB from Candida antarctica | | Descriptor: | 1,2-ETHANEDIOL, Lipase B | | Authors: | An, J, Xie, Y, Feng, Y, Wu, G. | | Deposit date: | 2013-04-15 | | Release date: | 2014-01-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhanced enzyme kinetic stability by increasing rigidity within the active site.
J.Biol.Chem., 289, 2014
|
|
3MYG
 
 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
5DWR
 
 | | Identification of N-(4-((1R,3S,5S)-3-amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1,2 and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies | | Descriptor: | N-{4-[(1R,3S,5S)-3-amino-5-methylcyclohexyl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-11 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies.
J.Med.Chem., 58, 2015
|
|
6O8N
 
 | |
6ILM
 
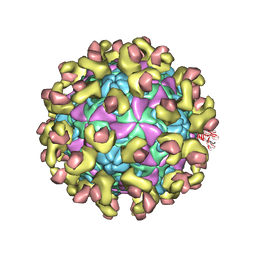 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 7.4 | | Descriptor: | Beta-2-microglobulin, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
4XRY
 
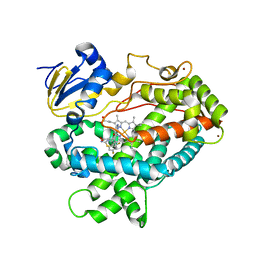 | | Human Cytochrome P450 2D6 BACE1 Inhibitor 5 Complex | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(1-methyl-1H-pyrazol-4-yl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Fan, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5ZXK
 
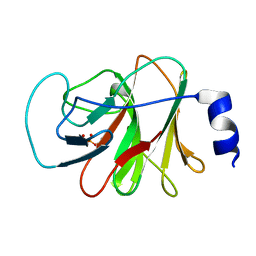 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6ILL
 
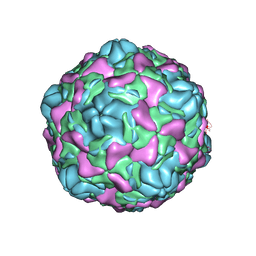 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6ILP
 
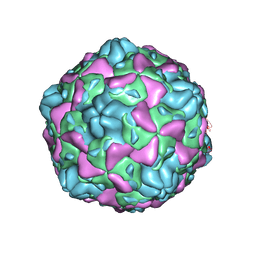 | | Cryo-EM structure of full Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6AEI
 
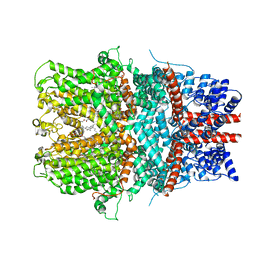 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-08-05 | | Release date: | 2019-08-07 | | Last modified: | 2019-08-14 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
6ILN
 
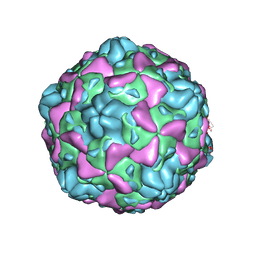 | | Cryo-EM structure of full Echovirus 6 particle at PH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
4XRZ
 
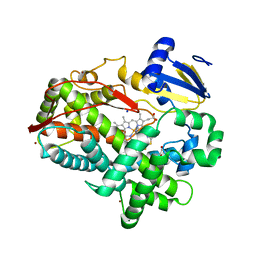 | | Human Cytochrome P450 2D6 BACE1 Inhibitor 6 Complex | | Descriptor: | (4aR,6R,8aS)-8a-(2,4-difluorophenyl)-6-(1H-pyrazol-4-yl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Cytochrome P450 2D6, GLYCEROL, ... | | Authors: | Johnson, E.F, Fan, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Utilizing Structures of CYP2D6 and BACE1 Complexes To Reduce Risk of Drug-Drug Interactions with a Novel Series of Centrally Efficacious BACE1 Inhibitors.
J.Med.Chem., 58, 2015
|
|
4MMJ
 
 | | crystal structure of YafQ from E.coli strain BL21(DE3) | | Descriptor: | Addiction module toxin, RelE/StbE family, SULFATE ION | | Authors: | Liang, Y, Gao, Z, Liu, Q, Dong, Y. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
1U6T
 
 | | Crystal structure of the human SH3 binding glutamic-rich protein like | | Descriptor: | CITRIC ACID, SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Yin, L, Xiang, Y, Yang, N, Zhu, D.-Y, Huang, R.-H, Wang, D.-C. | | Deposit date: | 2004-08-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human SH3BGRL protein: the first structure of the human SH3BGR family representing a novel class of thioredoxin fold proteins
Proteins, 61, 2005
|
|
6M71
 
 | | SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-03-16 | | Release date: | 2020-04-01 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
4N6Z
 
 | | Pim1 Complexed with a pyridylcarboxamide | | Descriptor: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | Deposit date: | 2013-10-14 | | Release date: | 2013-11-06 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
8D36
 
 | |
