2LN8
 
 | | The solution structure of theromacin | | Descriptor: | Theromacin | | Authors: | Jung, S, Soennichsen, F.D, Hung, C.-W, Tholey, A, Boidin-Wichlacz, C, Hausgen, W, Gelhaus, C, Desel, C, Podschun, R, Watzig, V, Tasiemski, A, Leippe, M, Groetzinger, J. | | Deposit date: | 2011-12-20 | | Release date: | 2012-03-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Macin family of antimicrobial proteins combines antimicrobial and nerve repair activities.
J.Biol.Chem., 287, 2012
|
|
5SZB
 
 | | Structure of human Dpf3 double-PHD domain bound to histone H3 tail peptide with acetylated K14 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Histone H3 tail peptide, ZINC ION, ... | | Authors: | Singh, N, Local, A, Shiau, A, Ren, B, Corbett, K.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification of H3K4me1-associated proteins at mammalian enhancers.
Nat. Genet., 50, 2018
|
|
1DF3
 
 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
2Y3Z
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - apo enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-ISOPROPYLMALATE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2LHL
 
 | | Chemical Shift Assignments and solution structure of human apo-S100A1 E32Q mutant | | Descriptor: | Protein S100-A1 | | Authors: | Ruszczynska-Bartnik, K, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N NMR sequence-specific resonance assignments and relaxation parameters for human apo-S100A1 E32Q mutant
To be Published
|
|
2Y40
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
1DK5
 
 | | CRYSTAL STRUCTURE OF ANNEXIN 24(CA32) FROM CAPSICUM ANNUUM | | Descriptor: | ANNEXIN 24(CA32), SULFATE ION | | Authors: | Hofmann, A, Proust, J, Dorowski, A, Schantz, R, Huber, R. | | Deposit date: | 1999-12-06 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Annexin 24 from Capsicum annuum. X-ray structure and biochemical characterization.
J.Biol.Chem., 275, 2000
|
|
1DDL
 
 | | DESMODIUM YELLOW MOTTLE TYMOVIRUS | | Descriptor: | DESMODIUM YELLOW MOTTLE VIRUS, RNA (5'-R(P*UP*U)-3'), RNA (5'-R(P*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Larson, S.B, Day, J, Canady, M.A, Greenwood, A, McPherson, A. | | Deposit date: | 1999-11-10 | | Release date: | 2000-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Refined structure of desmodium yellow mottle tymovirus at 2.7 A resolution.
J.Mol.Biol., 301, 2000
|
|
3FCJ
 
 | | Nitroalkane oxidase: mutant402N crystallized with nitroethane | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Nitroalkane oxidase, ... | | Authors: | Major, D.T, Gao, J, Heroux, A, Orville, A.M, Valley, M.P, Fitzpatrick, P.F. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Differential quantum tunneling contributions in nitroalkane oxidase catalyzed and the uncatalyzed proton transfer reaction.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5T2S
 
 | | Structure of the FHA1 domain of Rad53 bound simultaneously to the BRCT domain of Dbf4 and a phosphopeptide. | | Descriptor: | ASP-GLY-GLU-SER-TPO-ASP-GLU-ASP-ASP, DDK kinase regulatory subunit DBF4,Serine/threonine-protein kinase RAD53, GLYCEROL | | Authors: | Guarne, A, Almawi, A, Matthews, L. | | Deposit date: | 2016-08-24 | | Release date: | 2016-10-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 'AND' logic gates at work: Crystal structure of Rad53 bound to Dbf4 and Cdc7.
Sci Rep, 6, 2016
|
|
3TTG
 
 | | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum | | Descriptor: | CHLORIDE ION, Putative aminomethyltransferase | | Authors: | Michalska, K, Xu, X, Cui, H, Chin, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative aminomethyltransferase from Leptospirillum rubarum
TO BE PUBLISHED
|
|
2HWJ
 
 | | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu1540 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-01 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of protein Atu1540 from Agrobacterium tumefaciens
To be Published
|
|
1NRS
 
 | |
4J3I
 
 | | X-ray crystal structure of bromodomain complex to 1.24 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
4IWO
 
 | | Crystal structure and mechanism of activation of TBK1 | | Descriptor: | N-{3-[(5-cyclopropyl-2-{[3-(2-oxopyrrolidin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]propyl}cyclobutanecarboxamide, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Larabi, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure and mechanism of activation of TANK-binding kinase 1.
Cell Rep, 3, 2013
|
|
3FGT
 
 | | Two chain form of the 66.3 kDa protein from mouse lacking the linker peptide | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2008-12-08 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography
Bmc Struct.Biol., 9, 2009
|
|
4IYE
 
 | | Crystal structure of AdTx1 (rho-Da1a) from eastern green mamba (Dendroaspis angusticeps) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Toxin AdTx1 | | Authors: | Stura, E.A, Vera, L, Maiga, A.A, Marchetti, C, Lorphelin, A, Bellanger, L, Servant, D, Gilles, N. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystallization of recombinant green mamba rho-Da1a toxin during a lyophilization procedure and its structure determination.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2I1Y
 
 | | Crystal structure of the phosphatase domain of human PTP IA-2 | | Descriptor: | GLYCEROL, Receptor-type tyrosine-protein phosphatase | | Authors: | Faber-Barata, J, Patskovsky, Y, Alvarado, J, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Maletic, M, Rooney, I, Bain, K.T, Freeman, M, Russell, J.C, Thompson, D.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-15 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural genomics of protein phosphatases
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2QQU
 
 | | Crystal structure of a cell-wall invertase (D239A) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, ... | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Rabijns, A, Van den Ende, W. | | Deposit date: | 2007-07-27 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of Arabidopsis thaliana cell-wall invertase mutants in complex with sucrose.
J.Mol.Biol., 377, 2008
|
|
4J2K
 
 | | Crystal structure of a plant trypsin inhibitor EcTI | | Descriptor: | GLYCEROL, IMIDAZOLE, Trypsin inhibitor | | Authors: | Zhou, D, Wlodawer, A. | | Deposit date: | 2013-02-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of a Plant Trypsin Inhibitor from Enterolobium contortisiliquum (EcTI) and of Its Complex with Bovine Trypsin.
Plos One, 8, 2013
|
|
3FFP
 
 | | X ray structure of the complex between carbonic anhydrase II and LC inhibitors | | Descriptor: | 2-(1,3-thiazol-4-yl)-1H-benzimidazole-5-sulfonamide, BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Temperini, C, Crocetti, L, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A thiabendazole sulfonamide shows potent inhibitory activity against mammalian and nematode alpha-carbonic anhydrases
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2HY3
 
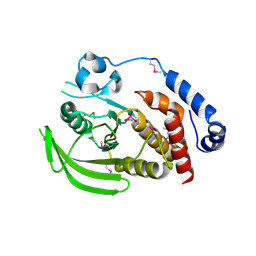 | | Crystal structure of the human tyrosine receptor phosphate gamma in complex with vanadate | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma, VANADATE ION | | Authors: | Jin, X, Min, T, Bera, A, Mu, H, Sauder, J.M, Freeman, J.C, Reyes, C, Smith, D, Wasserman, S.R, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3GSZ
 
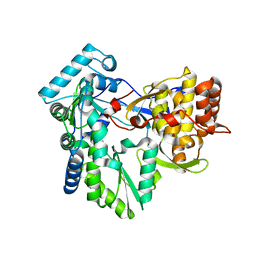 | | Structure of the genotype 2B HCV polymerase | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Rydberg, E.H, Carfi, A. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for resistance of the genotype 2b hepatitis C virus NS5B polymerase to site A non-nucleoside inhibitors.
J.Mol.Biol., 390, 2009
|
|
3UGH
 
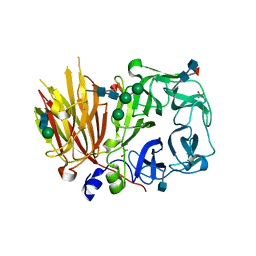 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
1PKR
 
 | |
