1FNZ
 
 | | A bark lectin from robinia pseudoacacia in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, BARK AGGLUTININ I, POLYPEPTIDE A, ... | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2000-08-24 | | Release date: | 2001-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine
Proteins, 44, 2001
|
|
1FNY
 
 | | LEGUME LECTIN OF THE BARK OF ROBINIA PSEUDOACACIA. | | Descriptor: | BARK AGGLUTININ I,POLYPEPTIDE A, CALCIUM ION | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2000-08-24 | | Release date: | 2001-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine.
Proteins, 44, 2001
|
|
1JY5
 
 | | RNase-related protein from Calystegia sepium | | Descriptor: | CalsepRRP | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2001-09-11 | | Release date: | 2002-04-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of an RNase-related protein from Calystegia sepium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1XXR
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus Nigra in complex with mannose | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
1XXQ
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus nigra | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
2SAK
 
 | | STAPHYLOKINASE (SAKSTAR VARIANT) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, STAPHYLOKINASE | | Authors: | Rabijns, A, De Bondt, H.L, De Maeyer, M, Lasters, I, De Ranter, C. | | Deposit date: | 1997-02-20 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of staphylokinase, a plasminogen activator with therapeutic potential.
Nat.Struct.Biol., 4, 1997
|
|
2Z79
 
 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
3HD8
 
 | | Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase | | Descriptor: | Endo-1,4-beta-xylanase A, Xylanase inhibitor | | Authors: | Sansen, S, Pollet, A, Raedschelders, G, Gebruers, K, Rabijns, A, Courtin, C.M. | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA
Febs J., 276, 2009
|
|
1R76
 
 | | Structure of a pectate lyase from Azospirillum irakense | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Novoa de Armas, H, Verboven, C, De Ranter, C, Desair, J, Vande Broek, A, Vanderleyden, J, Rabijns, A. | | Deposit date: | 2003-10-20 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Azospirillum irakense pectate lyase displays a toroidal fold.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MA9
 
 | | Crystal structure of the complex of human vitamin D binding protein and rabbit muscle actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Alpha Skeletal Muscle, ... | | Authors: | Verboven, C, Bogaerts, I, Waelkens, E, Rabijns, A, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2002-08-02 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Actin-DBP: the perfect structural fit?
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NNL
 
 | | Crystal structure of Human Phosphoserine Phosphatase | | Descriptor: | CALCIUM ION, CHLORIDE ION, L-3-phosphoserine phosphatase | | Authors: | Peeraer, Y, Rabijns, A, Verboven, C, Collet, J.F, Van Schaftingen, E, De Ranter, C. | | Deposit date: | 2003-01-14 | | Release date: | 2003-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of human phosphoserine phosphatase in open conformation.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3UGG
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 1-kestose | | Descriptor: | GLYCEROL, SULFATE ION, Sucrose:(Sucrose/fructan) 6-fructosyltransferase, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UGF
 
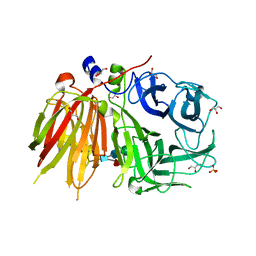 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
3UGH
 
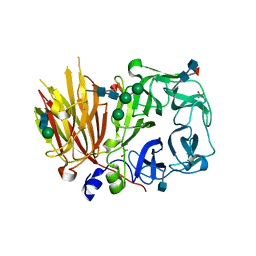 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 6-kestose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
1J7E
 
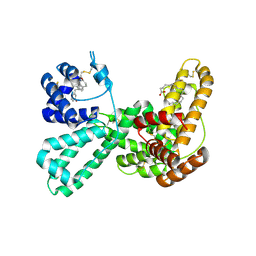 | | A Structural Basis for the Unique Binding Features of the Human Vitamin D-binding Protein | | Descriptor: | 3-(2-{4-[2-(5-HYDROXY-2-METHYLENE-CYCLOHEXYLIDENE)-ETHYLIDENE]-7A-METHYL-OCTAHYDRO-INDEN-1-YL}-PROPYL)-PHENOL, OLEIC ACID, vitamin D binding protein | | Authors: | Verboven, C, Rabijns, A, De Maeyer, M, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2001-05-16 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A structural basis for the unique binding features of the human vitamin D-binding protein.
Nat.Struct.Biol., 9, 2002
|
|
1J78
 
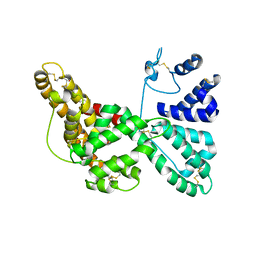 | | Crystallographic analysis of the human vitamin D binding protein | | Descriptor: | 3-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANOL, OLEIC ACID, vitamin D binding protein | | Authors: | Verboven, C, Rabijns, A, De Maeyer, M, Van Baelen, H, Bouillon, R, De Ranter, C. | | Deposit date: | 2001-05-16 | | Release date: | 2002-02-06 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural basis for the unique binding features of the human vitamin D-binding protein.
Nat.Struct.Biol., 9, 2002
|
|
1ST8
 
 | | Crystal structure of fructan 1-exohydrolase IIa from Cichorium intybus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Verhaest, M, Van den Ende, W, De Ranter, C.J, Van Laere, A, Rabijns, A. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | X-ray diffraction structure of a plant glycosyl hydrolase family 32 protein: fructan 1-exohydrolase IIa of Cichorium intybus.
Plant J., 41, 2005
|
|
1T6G
 
 | | Crystal structure of the Triticum aestivum xylanase inhibitor-I in complex with aspergillus niger xylanase-I | | Descriptor: | Endo-1,4-beta-xylanase I, GLYCEROL, xylanase inhibitor | | Authors: | Sansen, S, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
J.Biol.Chem., 279, 2004
|
|
1T6E
 
 | | Crystal Structure of the Triticum aestivum xylanase inhibitor I | | Descriptor: | GLYCEROL, xylanase inhibitor | | Authors: | Sansen, S, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
J.Biol.Chem., 279, 2004
|
|
3C7E
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7F
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7H
 
 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7G
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3EOX
 
 | |
