8KHU
 
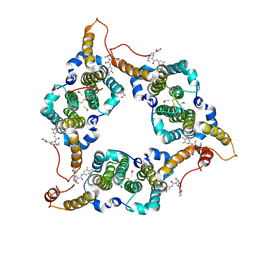 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
4O1I
 
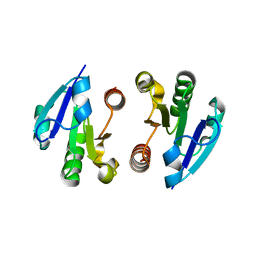 | | Crystal Structure of the regulatory domain of MtbGlnR | | Descriptor: | Transcriptional regulatory protein | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4O1H
 
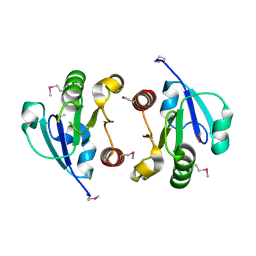 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
7XC2
 
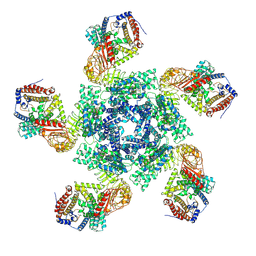 | | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Avirulence factor, CNL9 | | Authors: | Alexander, F, Li, E.T, Aaron, L, Deng, Y.N, Sun, Y, Chai, J.J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-21 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A wheat resistosome defines common principles of immune receptor channels.
Nature, 610, 2022
|
|
8AAA
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
5WXB
 
 | |
7DFV
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7WWM
 
 | | S protein of Delta variant in complex with ZWC6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7WWL
 
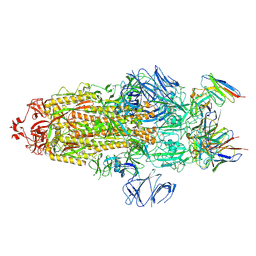 | | S protein of Delta variant in complex with ZWD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
1P5J
 
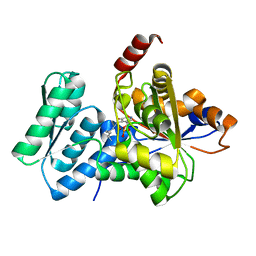 | | Crystal Structure Analysis of Human Serine Dehydratase | | Descriptor: | L-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sun, L, Liu, Y, Rao, Z. | | Deposit date: | 2003-04-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallization and preliminary crystallographic analysis of human serine dehydratase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7RTB
 
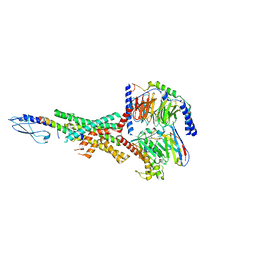 | | Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Johnson, R.M, Danev, R, Sexton, P.M, Wootten, D. | | Deposit date: | 2021-08-12 | | Release date: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
7S0V
 
 | |
5DEP
 
 | | Structure of Pseudomonas aeruginosa LpxA in complex with UDP-GlcNAc | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of Pseudomonas aeruginosa LpxA Reveal the Basis for Its Substrate Selectivity.
Biochemistry, 54, 2015
|
|
5DG3
 
 | |
5ZHE
 
 | | STRUCTURE OF E. COLI UNDECAPRENYL DIPHOSPHATE SYNTHASE IN COMPLEX WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
7DDZ
 
 | | The Crystal Structure of Human Neuropeptide Y Y2 Receptor with JNJ-31020028 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Human Neuropeptide Y Y2 Receptor fusion protein, ~{N}-[4-[4-[(1~{S})-2-(diethylamino)-2-oxidanylidene-1-phenyl-ethyl]piperazin-1-yl]-3-fluoranyl-phenyl]-2-pyridin-3-yl-benzamide | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2020-10-30 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for ligand recognition of the neuropeptide Y Y 2 receptor.
Nat Commun, 12, 2021
|
|
8ZAX
 
 | | Crystal structure of a short-chain dehydrogenase from Lactobacillus fermentum with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SDR family oxidoreductase | | Authors: | Wang, J.J, Cong, L, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-04-25 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
8GQ6
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8YAV
 
 | | Crystal structure of glucose 1-dehydrogenase from Limosilactobacillus fermentum | | Descriptor: | MAGNESIUM ION, SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-10 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir.
Adv.Synth.Catal., n/a, 2024
|
|
8YAI
 
 | | Crystal structure of glucose 1-dehydrogenase mutant1 from Limosilactobacillus fermentum | | Descriptor: | SDR family oxidoreductase | | Authors: | Cong, L, Wang, J.J, Wei, H.L, Liu, W.D, You, S. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Guided Engineering of a Short-Chain Dehydrogenase LfSDR1 for Efficient Biosynthesis of (R)-9-(2-Hydroxypropyl)adenine, the Key Intermediate of Tenofovir
Adv.Synth.Catal., 2024
|
|
8YAU
 
 | |
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
