1NL6
 
 | |
1NLJ
 
 | |
5TD9
 
 | | Structure of Human Enolase 2 | | Descriptor: | CHLORIDE ION, Gamma-enolase, MAGNESIUM ION | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2016-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.318 Å) | | Cite: | Pomhex, a cell-permeable Enolase inhibitor for Collateral Lethality targeting of ENO1-deleted Glioblastoma
To Be Published
|
|
7OSD
 
 | |
7OS8
 
 | | NMR SOLUTION STRUCTURE OF [Pro3,DLeu9]TL | | Descriptor: | PHE-VAL-PRO-TRP-PHE-SER-LYS-PHE-DLE-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A. | | Deposit date: | 2021-06-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | First-in-Class Cyclic Temporin L Analogue: Design, Synthesis, and Antimicrobial Assessment.
J.Med.Chem., 64, 2021
|
|
6V5N
 
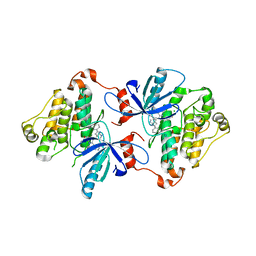 | | EGFR(T790M/V948R) in complex with LN2084 | | Descriptor: | 3-[4-(4-fluorophenyl)-5-(2-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)-1H-imidazol-2-yl]propan-1-ol, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V6O
 
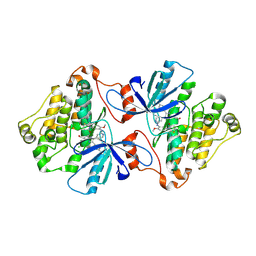 | | EGFR(T790M/V948R) in complex with LN2380 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(3-hydroxypropyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VHP
 
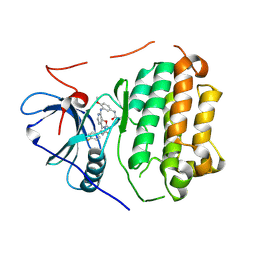 | | Wild type EGFR in complex with LN2899 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(4-{4-(4-fluorophenyl)-2-[(2-methoxyethyl)sulfanyl]-1H-imidazol-5-yl}pyridin-2-yl)amino]-4-methoxyphenyl}propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V6K
 
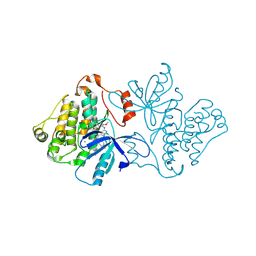 | | EGFR(T790M/V948R) in complex with LN2057 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(methylsulfanyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VH4
 
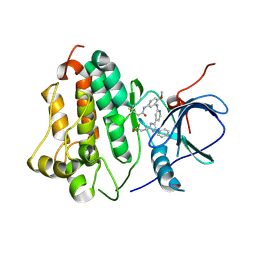 | | Wild type EGFR in complex with LN2380 | | Descriptor: | Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(3-hydroxypropyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V66
 
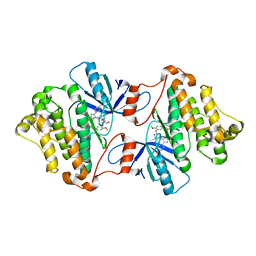 | | EGFR(T790M/V948R) in complex with LN2899 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Epidermal growth factor receptor, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6V5P
 
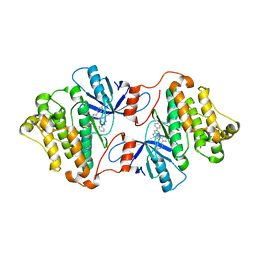 | | EGFR(T790M/V948R) in complex with LN2725 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-(3-methoxypropyl)-1H-imidazol-5-yl]-2-phenyl-3H-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VHN
 
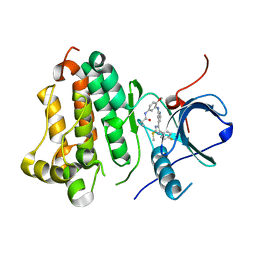 | | Wild type EGFR in complex with LN2057 | | Descriptor: | Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(methylsulfanyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6F4U
 
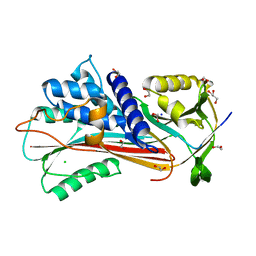 | |
7O4W
 
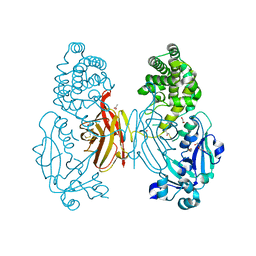 | |
7P40
 
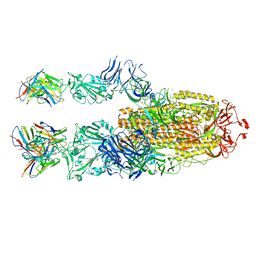 | | P5C3 is a potent fab neutralizer | | Descriptor: | Spike glycoprotein, Variable Heavy Chain P5C3 (VH), Variable Light Chain P5C3 (VL) | | Authors: | perez, L. | | Deposit date: | 2021-07-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7PHG
 
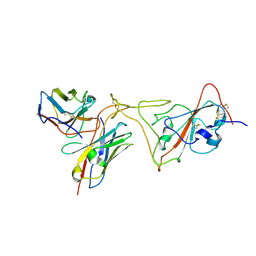 | | MaP OF P5C3RBD Interface | | Descriptor: | Heavy ChaIn variable, Light ChaIn, Surface glycoprotein | | Authors: | Perez, L. | | Deposit date: | 2021-08-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A highly potent antibody effective against SARS-CoV-2 variants of concern.
Cell Rep, 37, 2021
|
|
7MBI
 
 | | Structure of SARS-CoV2 3CL protease covalently bound to peptidomimetic inhibitor | | Descriptor: | 2,4,6-trimethylpyridine-3-carboxylic acid, 3C-like proteinase, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-3-oxo-1-[(3S)-2-oxopiperidin-3-yl]butan-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Khan, M.B, Lu, J, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Peptidomimetic alpha-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability.
J.Med.Chem., 65, 2022
|
|
1AGQ
 
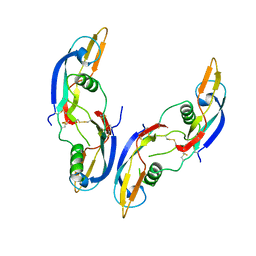 | | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR FROM RAT | | Descriptor: | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR | | Authors: | Eigenbrot, C, Gerber, N. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of glial cell-derived neurotrophic factor at 1.9 A resolution and implications for receptor binding.
Nat.Struct.Biol., 4, 1997
|
|
5QII
 
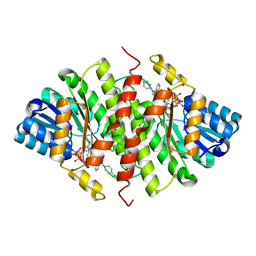 | | CRYSTAL STRUCTURE OF 11BETA-HSD1 DOUBLE MUTANT (L262R, F278E) COMPLEXED WITH 2-(3-(1-(4-CHLOROPHENYL)CYCLOPROPYL) -[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8-YL)PROPAN-2-OL | | Descriptor: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sheriff, S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
5QIJ
 
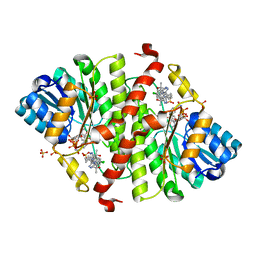 | | CRYSTAL STRUCTURE OF MURINE 11B- HYDROXYSTEROIDDEHYDROGENASE COMPLEXED WITH 2-(3-(1-(4- CHLOROPHENYL)CYCLOPROPYL)-[1,2,4]TRIAZOLO[4,3-A]PYRIDIN-8- YL)PROPAN-2-OL | | Descriptor: | 2-{3-[1-(4-chlorophenyl)cyclopropyl][1,2,4]triazolo[4,3-a]pyridin-8-yl}propan-2-ol, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of Clinical Candidate BMS-823778 as an Inhibitor of Human 11 beta-Hydroxysteroid Dehydrogenase Type 1 (11 beta-HSD-1).
ACS Med Chem Lett, 9, 2018
|
|
2H8B
 
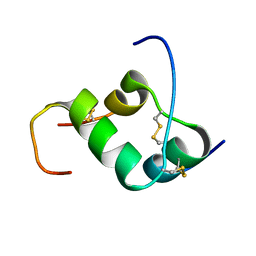 | | Solution structure of INSL3 | | Descriptor: | Insulin-like 3 | | Authors: | Rosengren, K.J, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Characterization of the LGR8 Receptor Binding Surface of Insulin-like Peptide 3
J.Biol.Chem., 281, 2006
|
|
2FHW
 
 | | Solution structure of human relaxin-3 | | Descriptor: | Relaxin 3 (Prorelaxin H3) (Insulin-like peptide INSL7) (Insulin-like peptide 7) | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2005-12-27 | | Release date: | 2006-01-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure and novel insights into the determinants of the receptor specificity of human relaxin-3.
J.Biol.Chem., 281, 2006
|
|
7OSE
 
 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
5K27
 
 | | Crystal structure of ancestral protein ancMT of ADP-dependent sugar kinases family. | | Descriptor: | ADENOSINE MONOPHOSPHATE, IODIDE ION, ancMT | | Authors: | Castro-Fernandez, V, Herrera-Morande, A, Zamora, R, Merino, F, Pereira, H.M, Brandao-Neto, J, Garratt, R, Guixe, V. | | Deposit date: | 2016-05-18 | | Release date: | 2017-05-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Reconstructed ancestral enzymes reveal that negative selection drove the evolution of substrate specificity in ADP-dependent kinases.
J. Biol. Chem., 292, 2017
|
|
