3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | Descriptor: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNK
 
 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | Descriptor: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNI
 
 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | Descriptor: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
2MI2
 
 | |
2M3I
 
 | | Characterization of a Novel Alpha4/6-Conotoxin TxIC from Conus textile that Potently Blocks alpha3beta4 Nicotinic Acetylcholine Receptors | | Descriptor: | Alpha-conotoxin | | Authors: | Luo, S, Zhangsun, D, Zhu, X, Wu, Y, Hu, Y, Christensen, S, Akcan, M, Craik, D.J, McIntosh, J.M. | | Deposit date: | 2013-01-20 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 56, 2013
|
|
2LZ5
 
 | | Solution structure of a Novel Alpha-Conotoxin TxIB | | Descriptor: | Conotoxin_TxIB | | Authors: | Luo, S, Zhangsun, D, Wu, Y, Zhu, X, Hu, Y, McIntyre, M, Christensen, S, Akcan, M, Craik, D, McIntosh, J. | | Deposit date: | 2012-09-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of a novel alpha-conotoxin from conus textile that selectively targets alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors.
J.Biol.Chem., 288, 2013
|
|
2MN6
 
 | |
2MN7
 
 | |
7T64
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T65
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | Deposit date: | 2021-12-13 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6UG0
 
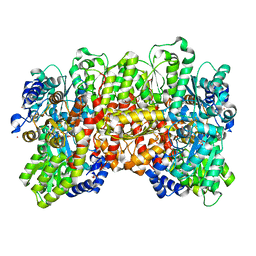 | | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2019-09-25 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
7JMA
 
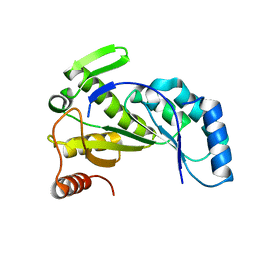 | |
7JMB
 
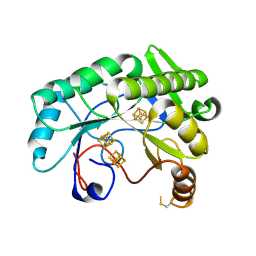 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | Authors: | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
6WOV
 
 | | Cryo-EM structure of recombinant mouse Ryanodine Receptor type 2 wild type in complex with FKBP12.6 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Iyer, K.A, Hu, Y, Nayak, A.R, Kurebayashi, N, Murayama, T, Samso, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
7MCI
 
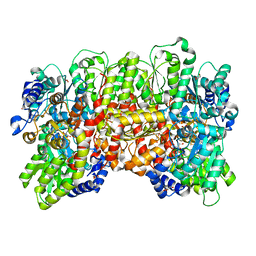 | | MoFe protein from Azotobacter vinelandii with a sulfur-replenished cofactor | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Lee, C, Hu, Y, Ribbe, M.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nat Catal, 5, 2022
|
|
3W02
 
 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W01
 
 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZZ
 
 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZX
 
 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W00
 
 | | Crystal structure of PcrB complexed with G1P and FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3WQK
 
 | | Crystal structure of Rv3378c with PO4 | | Descriptor: | Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3WQM
 
 | | Crystal structure of Rv3378c with inhibitor BPH-629 | | Descriptor: | Diterpene synthase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3VNC
 
 | | Crystal Structure of TIP-alpha N25 from Helicobacter Pylori in its natural dimeric form | | Descriptor: | TIP-alpha | | Authors: | Gao, M, Li, D, Hu, Y, Zou, Q, Wang, D.-C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of TNF-alpha-Inducing Protein from Helicobacter Pylori in Active Form Reveals the Intrinsic Molecular Flexibility for Unique DNA-Binding
Plos One, 7, 2012
|
|
