4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
2F8P
 
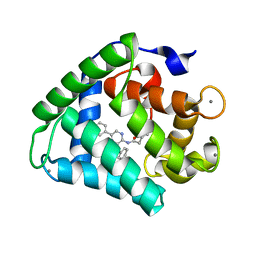 | | Crystal structure of obelin following Ca2+ triggered bioluminescence suggests neutral coelenteramide as the primary excited state | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TNO
 
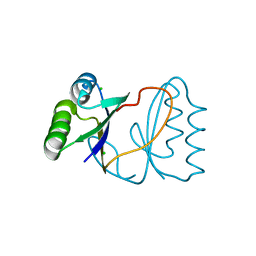 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
6B1B
 
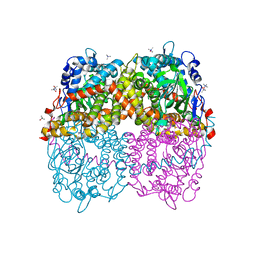 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI MUTANT XS6 (APO Enzyme) | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, trimethylamine oxide | | Authors: | Zhou, D, Kandavelu, P, Wang, B.C, Yan, Y, Rose, J.P. | | Deposit date: | 2017-09-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
1GSB
 
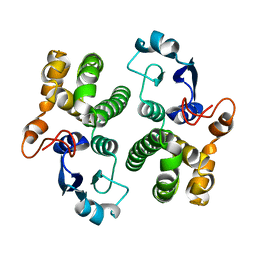 | |
1GSC
 
 | |
1JF2
 
 | | Crystal Structure of W92F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, obelin | | Authors: | Liu, Z.-J, Vysotski, E.S, Deng, L, Markova, S.V, Lee, J, Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-06-19 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Violet bioluminescence and fast kinetics from W92F obelin: structure-based proposals for the bioluminescence triggering and the identification of the emitting species.
Biochemistry, 42, 2003
|
|
3OV8
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus, High resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, Protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-09-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8501 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3O3K
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J.-Y, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Xu, H, Chen, L, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Archaeoglobus fulgidus orphan ORF AF1382 determined by sulfur SAD from a moderately diffracting crystal.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4LB2
 
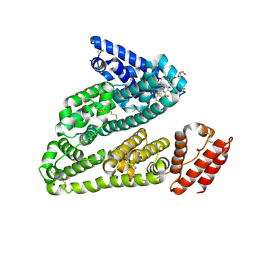 | | X-ray study of human serum albumin complexed with idarubicin | | Descriptor: | IDARUBICIN, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LB9
 
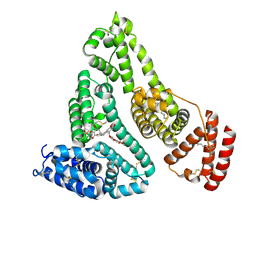 | | X-ray study of human serum albumin complexed with etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4L9K
 
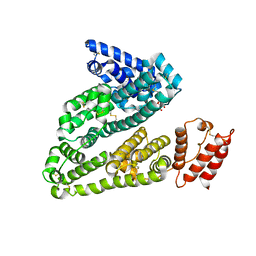 | | X-ray study of human serum albumin complexed with camptothecin | | Descriptor: | (2S)-2-hydroxy-2-[8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]butanoic acid, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4L8U
 
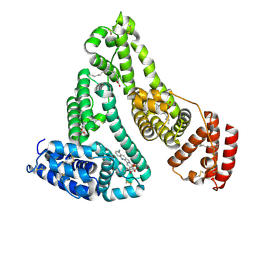 | | X-ray study of human serum albumin complexed with 9 amino camptothecin | | Descriptor: | (2S)-2-[1-amino-8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]-2-hydroxybutanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-17 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LA0
 
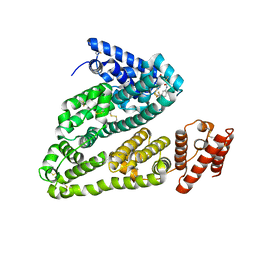 | | X-ray study of human serum albumin complexed with bicalutamide | | Descriptor: | R-BICALUTAMIDE, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4L9Q
 
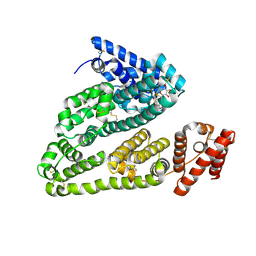 | | X-ray study of human serum albumin complexed with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
7N0J
 
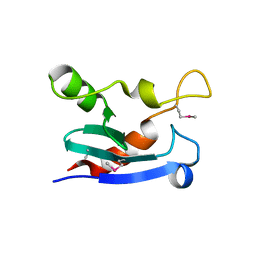 | | Structure of YebY from E. coli K12 | | Descriptor: | YebY | | Authors: | Hadley, R.C, Rosenzweig, A.C. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The copper-linked Escherichia coli AZY operon: Structure, metal binding, and a possible physiological role in copper delivery.
J.Biol.Chem., 298, 2022
|
|
4MN0
 
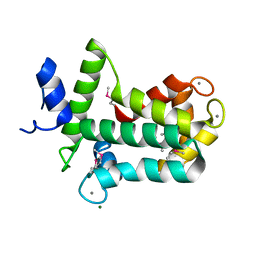 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | Descriptor: | Berovin, CALCIUM ION, MAGNESIUM ION | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
4R8U
 
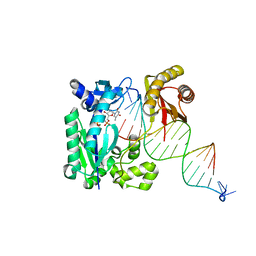 | | S-SAD structure of DINB-DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA, DNA polymerase IV, ... | | Authors: | Kottur, J, Nair, D.T, Weinert, T, Oligeric, V, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination
Nat.Methods, 12, 2015
|
|
4R8T
 
 | | Structure of JEV protease | | Descriptor: | CHLORIDE ION, NS3, Serine protease subunit NS2B | | Authors: | Nair, D.T, Weinert, T, Wang, M, Olieric, V. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
3SZB
 
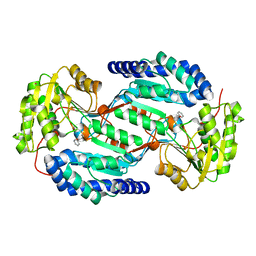 | |
3SZA
 
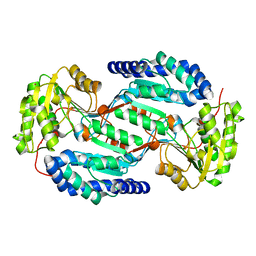 | | Crystal structure of human ALDH3A1 - apo form | | Descriptor: | ACETATE ION, Aldehyde dehydrogenase, dimeric NADP-preferring, ... | | Authors: | Khanna, M, Hurley, T.D. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases.
J.Biol.Chem., 286, 2011
|
|
5WI4
 
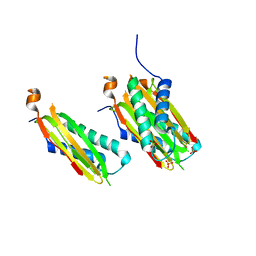 | | CRYSTAL STRUCTURE OF DYNLT1/TCTEX-1 IN COMPLEX WITH ARHGEF2 | | Descriptor: | Dynein light chain Tctex-type 1,Rho guanine nucleotide exchange factor 2, SULFATE ION | | Authors: | Balan, M, Ishiyama, N, Marshall, C.B, Ikura, M. | | Deposit date: | 2017-07-18 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity.
Sci Signal, 10, 2017
|
|
2HPV
 
 | | Crystal structure of FMN-Dependent azoreductase from Enterococcus faecalis | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Liu, Z.J, Chen, L, Chen, H, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fmn-Dependent Azoreductase from Enterococcus faecalis at 2.00 A resolution
To be Published
|
|
1LKK
 
 | |
