2FMX
 
 | | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+) | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rao, Y, Bian, C, Yuan, C, Li, Y, Huang, M. | | Deposit date: | 2006-01-10 | | Release date: | 2006-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg(2+)
Biochem.Biophys.Res.Commun., 348, 2006
|
|
2A0T
 
 | | NMR structure of the FHA1 domain of Rad53 in complex with a biological relevant phosphopeptide derived from Madt1 | | Descriptor: | Hypothetical 73.8 kDa protein in SAS3-SEC17 intergenic region, residues 301-310, Serine/threonine-protein kinase RAD53 | | Authors: | Mahajan, A, Yuan, C, Pike, B.L, Heierhorst, J, Chang, C.-F, Tsai, M.-D. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | FHA Domain-Ligand Interactions: Importance of Integrating Chemical and Biological Approaches
J.Am.Chem.Soc., 127, 2005
|
|
4XSK
 
 | | Structure of PAItrap, an uPA mutant | | Descriptor: | GLYCEROL, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L, Proulle, V, Hong, Z, Lin, Z, Liu, M, Yuan, C, Lin, L, Furie, B, Flaumenhaft, R, Andreasen, P, Furie, B, Huang, M. | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PAItrap, an uPA mutant
To Be Published
|
|
3TKZ
 
 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | Descriptor: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
3TL0
 
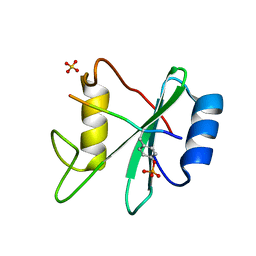 | | Structure of SHP2 N-SH2 domain in complex with RLNpYAQLWHR peptide | | Descriptor: | RLNpYAQLWHR peptide, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
4QTH
 
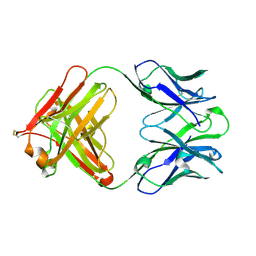 | | Crystal structure of anti-uPAR Fab 8B12 | | Descriptor: | anti-uPAR antibody, heavy chain, light chain | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
4QTI
 
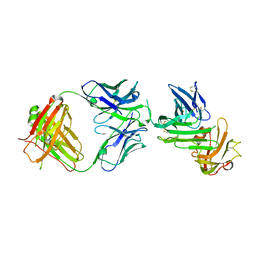 | | Crystal structure of human uPAR in complex with anti-uPAR Fab 8B12 | | Descriptor: | Urokinase plasminogen activator surface receptor, anti-uPAR antibody, heavy chain, ... | | Authors: | Zhao, B, Yuan, C, Luo, Z, Huang, M. | | Deposit date: | 2014-07-08 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stabilizing a flexible interdomain hinge region harboring the SMB binding site drives uPAR into its closed conformation.
J.Mol.Biol., 427, 2015
|
|
3G7N
 
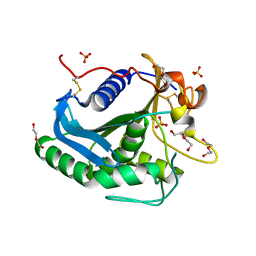 | | Crystal Structure of a Triacylglycerol Lipase from Penicillium Expansum at 1.3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lipase, PENTAETHYLENE GLYCOL, ... | | Authors: | Bian, C.B, Yuan, C, Chen, L.Q, Edward, J.M, Lin, L, Jiang, L.G, Huang, Z.X, Huang, M.D. | | Deposit date: | 2009-02-10 | | Release date: | 2010-02-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a triacylglycerol lipase from Penicillium expansum at 1.3 A determined by sulfur SAD
Proteins, 78, 2010
|
|
4DVA
 
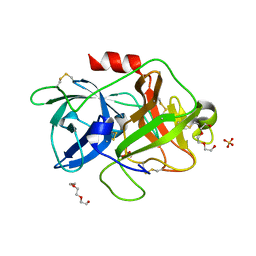 | | The crystal structure of human urokinase-type plasminogen activator catalytic domain | | Descriptor: | HEXAETHYLENE GLYCOL, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody
Biochem.J., 449, 2013
|
|
4DVB
 
 | | The crystal structure of the Fab fragment of pro-uPA antibody mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
4DW2
 
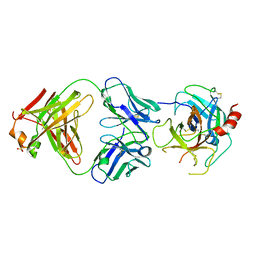 | | The crystal structure of uPA in complex with the Fab fragment of mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
8Y7F
 
 | |
8Y6Z
 
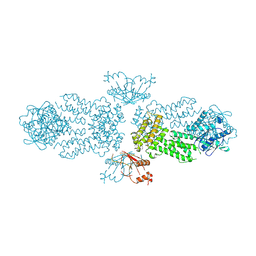 | |
8Y7G
 
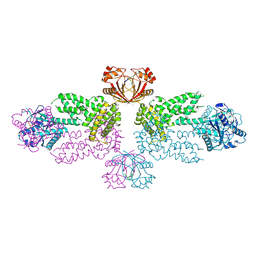 | |
8Y75
 
 | |
2N9C
 
 | | NRAS Isoform 5 | | Descriptor: | GTPase NRas | | Authors: | Markowitz, J, Mal, T.K, Yuan, C, Courtney, N.B, Patel, M, Stiff, A.R, Blachly, J, Walker, C, Eisfeld, A, de la Chapelle, A, Carson III, W.E. | | Deposit date: | 2015-11-13 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of NRAS isoform 5.
Protein Sci., 25, 2016
|
|
3U73
 
 | | Crystal structure of stabilized human uPAR mutant in complex with ATF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator, ... | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of the urokinase receptor in a ligand-free form.
J.Mol.Biol., 416, 2012
|
|
3U74
 
 | | Crystal structure of stabilized human uPAR mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the urokinase receptor in a ligand-free form.
J.Mol.Biol., 416, 2012
|
|
3KHV
 
 | | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol | | Descriptor: | SULFATE ION, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator, ... | | Authors: | Jiang, L.-G, Zhao, G.-X, Bian, X.-B, Yuan, C, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3KID
 
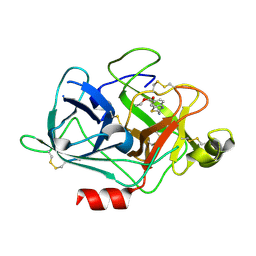 | | The Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator | | Descriptor: | Urokinase-type plasminogen activator, ethyl 2-amino-1,3-benzothiazole-6-carboxylate | | Authors: | Jiang, L.-G, Yu, H.Y, Yuan, C, Wang, J.D, Chen, L.Q, Meehan, E.J, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-11-01 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3KGP
 
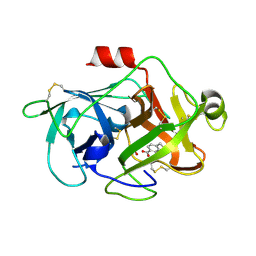 | | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol | | Descriptor: | 4-(aminomethyl)benzoic acid, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Zhao, G.-X, Bian, X.-B, Yuan, C, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Urokinase-type Plasminogen Activator in Complex with 4-(Aminomethyl) Benzoic Acid and 4-(Aminomethyl-phenyl)-methanol
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3HNB
 
 | |
3HNY
 
 | |
3HOB
 
 | |
2JXJ
 
 | |
