2XYV
 
 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYQ
 
 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYR
 
 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
3OV9
 
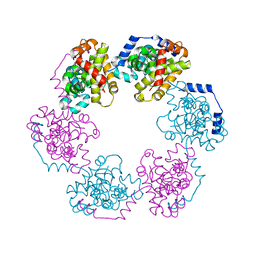 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein, SODIUM ION | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
3OUO
 
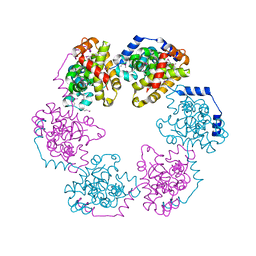 | | Structure of the Nucleoprotein from Rift Valley Fever Virus | | Descriptor: | NITRITE ION, Nucleoprotein | | Authors: | Ferron, F, Danek, E.I, Li, Z, Luo, D, Wong, Y.H, Coutard, B, Lantez, V, Charrel, R, Canard, B, Walz, T, Lescar, J. | | Deposit date: | 2010-09-15 | | Release date: | 2011-05-25 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes
Plos Pathog., 7, 2011
|
|
3Q3X
 
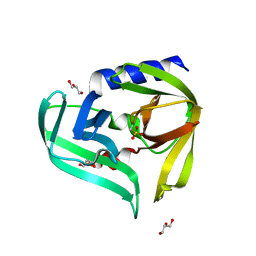 | | Crystal structure of the main protease (3C) from human enterovirus B EV93 | | Descriptor: | GLYCEROL, HEVB EV93 3C protease, MAGNESIUM ION | | Authors: | Costenaro, L, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3Q3Y
 
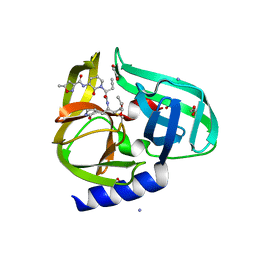 | | Complex structure of HEVB EV93 main protease 3C with Compound 1 (AG7404) | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, HEVB EV93 3C protease, ... | | Authors: | Costenaro, L, Kaczmarska, Z, Arnan, C, Sola, M, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2010-12-22 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3RUO
 
 | | Complex structure of HevB EV93 main protease 3C with Rupintrivir (AG7088) | | Descriptor: | 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, CHLORIDE ION, HEVB EV93 3C PROTEASE, ... | | Authors: | Kaczmarska, Z, Janowski, R, Costenaro, L, Coutard, B, Norder, H, Canard, B, Coll, M. | | Deposit date: | 2011-05-05 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Antiviral Inhibition of the Main Protease, 3C, from Human Enterovirus 93.
J.Virol., 85, 2011
|
|
3GZF
 
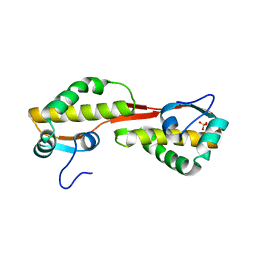 | | Structure of the C-terminal domain of nsp4 from Feline Coronavirus | | Descriptor: | Replicase polyprotein 1ab, SULFATE ION | | Authors: | Manolaridis, I, Wojdyla, J.A, Panjikar, S, Snijder, E.J, Gorbalenya, A.E, Coutard, B, Tucker, P.A. | | Deposit date: | 2009-04-07 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Structure of the C-terminal domain of nsp4 from feline coronavirus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5FVA
 
 | | Toscana Virus Nucleocapsid Protein | | Descriptor: | NUCLEOPROTEIN, SODIUM ION | | Authors: | Baklouti, A, Sevajol, M, Goulet, A, Lichiere, J, Ferron, F, Canard, B, Charrel, R.N, Coutard, B, Papageorgiou, N. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Toscana virus nucleoprotein oligomer organization observed in solution.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
3GPG
 
 | |
3GPO
 
 | | Crystal structure of macro domain of Chikungunya virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GQE
 
 | | Crystal structure of macro domain of Venezuelan Equine Encephalitis virus | | Descriptor: | BICINE, Non-structural protein 3 | | Authors: | Jamal, S, Malet, H, Coutard, B, Canard, B. | | Deposit date: | 2009-03-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GQO
 
 | | Crystal structure of macro domain of Venezuelan Equine Encephalitis virus in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Malet, H, Jamal, S, Coutard, B, Ferron, F, Canard, B. | | Deposit date: | 2009-03-24 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
3GPQ
 
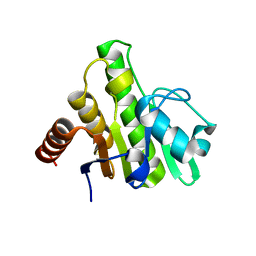 | | Crystal structure of macro domain of Chikungunya virus in complex with RNA | | Descriptor: | Non-structural protein 3, RNA (5'-R(*AP*AP*A)-3') | | Authors: | Malet, H, Jamal, S, Coutard, B, Canard, B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket
J.Virol., 83, 2009
|
|
4CTK
 
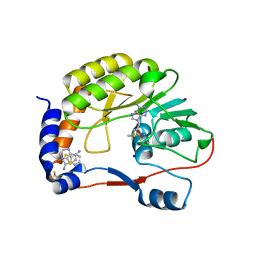 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYL METHIONINE AND FRAGMENT 2A4 | | Descriptor: | DIMETHYL SULFOXIDE, POLYPROTEIN, S-ADENOSYLMETHIONINE, ... | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2014-03-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Assessment of Dengue Virus Helicase and Methyltransferase as Targets for Fragment-Based Drug Discovery.
Antiviral Res., 106, 2014
|
|
4CTJ
 
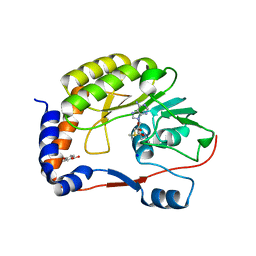 | | DENGUE 3 NS5 METHYLTRANSFERASE BOUND TO S-ADENOSYL METHIONINE AND FRAGMENT 3A9 | | Descriptor: | 2,3-dihydro-1-benzofuran-5-carboxylic acid, NON-STRUCTURAL PROTEIN 5, S-ADENOSYLMETHIONINE, ... | | Authors: | Barral, K, Bricogne, G, Sharff, A. | | Deposit date: | 2014-03-14 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Assessment of Dengue Virus Helicase and Methyltransferase as Targets for Fragment-Based Drug Discovery.
Antiviral Res., 106, 2014
|
|
4Y2A
 
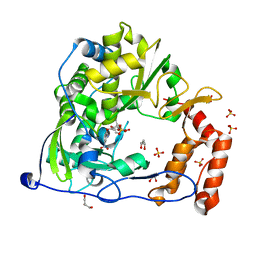 | | Crystal Structure of Coxsackie Virus B3 3D polymerase in complex with GPC-N114 inhibitor | | Descriptor: | 2,2'-[(4-chlorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | Authors: | Vives-Adrian, L, Ferrer-Orta, C, Verdaguer, N. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
6YU8
 
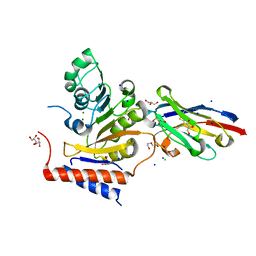 | | RNA Methyltransferase of Sudan Ebola Virus | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ferron, F, Valle, C, Zamboni, V, Canard, B, Decroly, E. | | Deposit date: | 2020-04-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | First insights into the structural features of Ebola virus methyltransferase activities.
Nucleic Acids Res., 49, 2021
|
|
4NZ0
 
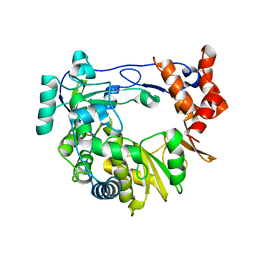 | |
4NYZ
 
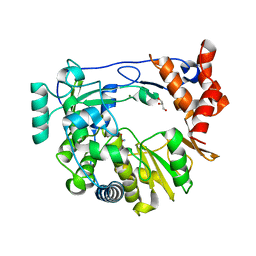 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | Descriptor: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | Authors: | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
4Y34
 
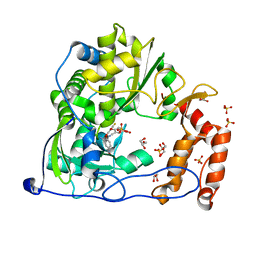 | | Crystal Structure of Coxsackievirus B3 3D polymerase in complex with GPC-N143 | | Descriptor: | 2,2'-[(4-fluorobenzene-1,2-diyl)bis(oxy)]bis(5-nitrobenzonitrile), 3D polymerase, GLYCEROL, ... | | Authors: | Vives-Adrian, L, Ferrer-Orta, C, Cerdaguer, N. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
4Y3C
 
 | | I304V 3D polymerase mutant of EMCV | | Descriptor: | 3D polymerase, CHLORIDE ION, GLYCEROL | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
4Y2C
 
 | | M300V 3D polymerase mutant of EMCV | | Descriptor: | GLYCEROL, Genome polyprotein | | Authors: | Verdaguer, N, Ferrer-Orta, C, Vives-Adrian, L. | | Deposit date: | 2015-02-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family.
Plos Pathog., 11, 2015
|
|
2QEQ
 
 | |
