2JTC
 
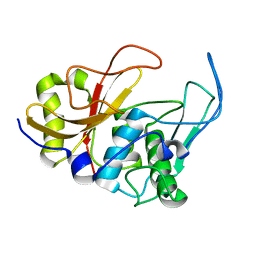 | | 3D structure and backbone dynamics of SPE B | | Descriptor: | Streptopain | | Authors: | Chuang, W, Wang, C, Houng, H, Chen, C, Wang, P. | | Deposit date: | 2007-07-26 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of streptopain: insight into diverse substrate specificity.
J.Biol.Chem., 284, 2009
|
|
4XZ6
 
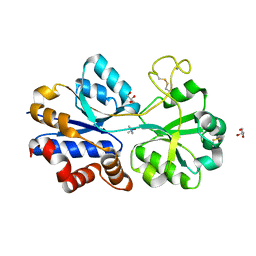 | | TmoX in complex with TMAO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic Insight into Trimethylamine N-Oxide Recognition by the Marine Bacterium Ruegeria pomeroyi DSS-3
J.Bacteriol., 197, 2015
|
|
3VV3
 
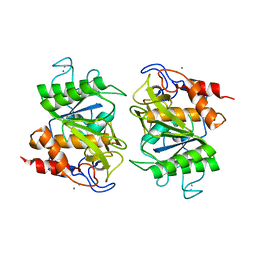 | | Crystal structure of deseasin MCP-01 from Pseudoalteromonas sp. SM9913 | | Descriptor: | CALCIUM ION, Deseasin MCP-01 | | Authors: | Zhao, G.Y, Gao, X, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2012-07-14 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into collagen degradation by a bacterial collagenolytic serine protease in the subtilisin family.
Mol. Microbiol., 90, 2013
|
|
7L2C
 
 | | Crystallographic structure of neutralizing antibody 2-51 in complex with SARS-CoV-2 spike N-terminal domain (NTD) | | Descriptor: | 2-51 heavy chain, 2-51 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cerutti, G, Reddem, E.R, Shapiro, L. | | Deposit date: | 2020-12-16 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
7L2E
 
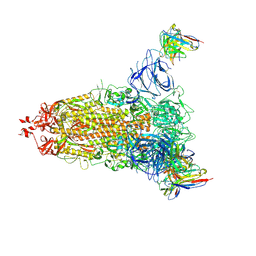 | |
7L2D
 
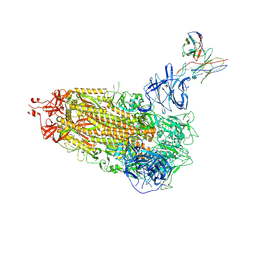 | |
7L2F
 
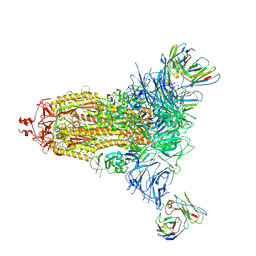 | |
7VS2
 
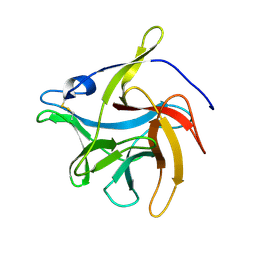 | | secreted fungal effector protein MoErs1 | | Descriptor: | MoErs1 | | Authors: | Wang, F.F, Xing, W.M. | | Deposit date: | 2021-10-25 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Magnaporthe oryzae effector MoErs1 and host papain-like protease OsRD21 interaction to combat rice blast.
Nat.Plants, 2024
|
|
7CQQ
 
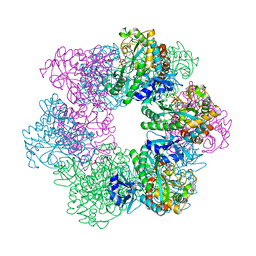 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7LQV
 
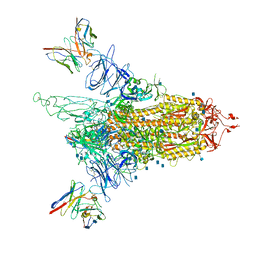 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7CQL
 
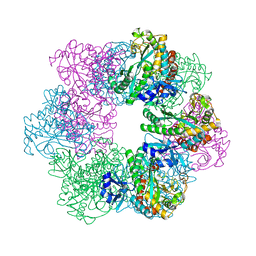 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
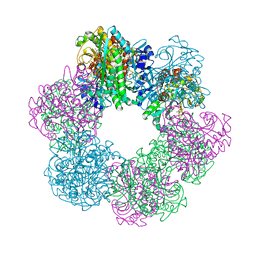 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
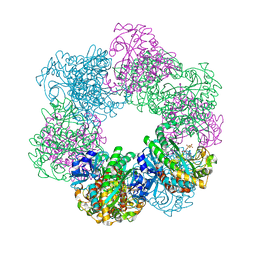 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
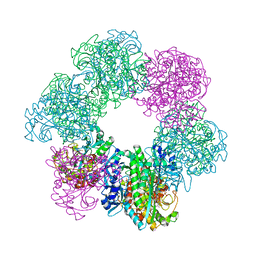 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CKM
 
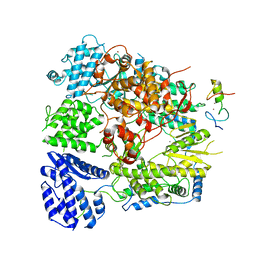 | | Structure of Machupo virus polymerase bound to Z matrix protein (monomeric complex) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7CKL
 
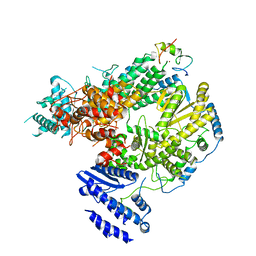 | | Structure of Lassa virus polymerase bound to Z matrix protein | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7CM9
 
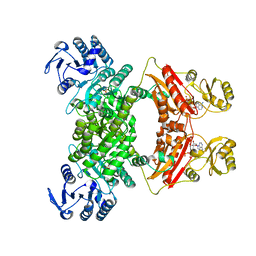 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
6BMV
 
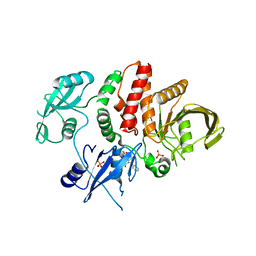 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP504 | | Descriptor: | 3-{4-[(2-chlorophenyl)methyl]-5-oxo-4,5-dihydro[1,2,4]triazolo[4,3-a]quinazolin-1-yl}-4-hydroxybenzoic acid, PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMU
 
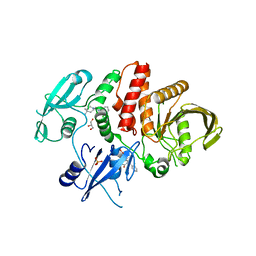 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP244 | | Descriptor: | 4-[(2-chlorophenyl)methyl]-1-(2-hydroxy-3-methoxyphenyl)[1,2,4]triazolo[4,3-a]quinazolin-5(4H)-one, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMR
 
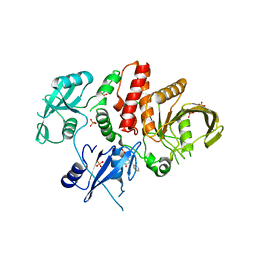 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP244 | | Descriptor: | 4-[(2-chlorophenyl)methyl]-1-(2-hydroxy-3-methoxyphenyl)[1,2,4]triazolo[4,3-a]quinazolin-5(4H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMX
 
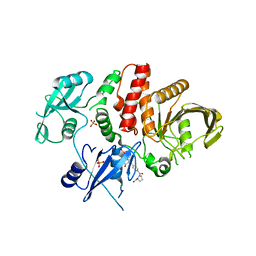 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP844 | | Descriptor: | 1-(3-chloro-4-{[1-(2-hydroxy-3-methoxyphenyl)-5-oxo[1,2,4]triazolo[4,3-a]quinazolin-4(5H)-yl]methyl}benzene-1-carbonyl)-L-proline, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6BMY
 
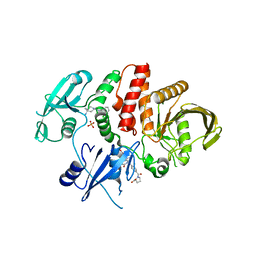 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitors SHP099 and SHP844 | | Descriptor: | 1-(3-chloro-4-{[1-(2-hydroxy-3-methoxyphenyl)-5-oxo[1,2,4]triazolo[4,3-a]quinazolin-4(5H)-yl]methyl}benzene-1-carbonyl)-L-proline, 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, GLYCEROL, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
7D4N
 
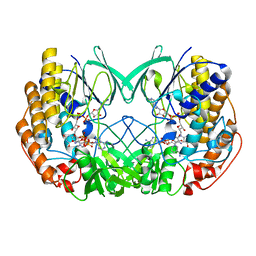 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 20 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4M
 
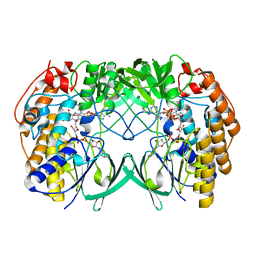 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 5 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
