6AA2
 
 | | X-ray structure of ReQy1 (oxidized form) | | Descriptor: | Green fluorescent protein | | Authors: | Sugiura, K, Yasuda, A, Tabushi, N, Tanaka, H, Kurisu, G, Hisabori, T. | | Deposit date: | 2018-07-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5ZZ4
 
 | | Crystal structure of bruton's tyrosine kinase in complex with inhibitor 2e | | Descriptor: | N-[3-(4-amino-6-{[4-(morpholine-4-carbonyl)phenyl]amino}-1,3,5-triazin-2-yl)-2-methylphenyl]-4-tert-butylbenzamide, Tyrosine-protein kinase BTK | | Authors: | Kawahata, W, Asami, T, Irie, T, Kiyoi, T, Taniguchi, H, Asamitsu, Y, Inoue, T, Miyake, T, Sawa, M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design and Synthesis of Novel Amino-triazine Analogues as Selective Bruton's Tyrosine Kinase Inhibitors for Treatment of Rheumatoid Arthritis.
J. Med. Chem., 61, 2018
|
|
6AA6
 
 | |
3W39
 
 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
3VIS
 
 | | Crystal structure of cutinase Est119 from Thermobifida alba AHK119 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Esterase | | Authors: | Kitadokoro, K, Thumarat, U, Nakamura, R, Nishimura, K, Karatani, H, Suzuki, H, Kawai, F. | | Deposit date: | 2011-10-11 | | Release date: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of cutinase Est119 from Thermobida alba AHK119 that can degrade modpolyethylene terephthalate at 1.76 A resolution.
POLYM.DEGRAD.STAB., 97, 2012
|
|
3WII
 
 | | Crystal structure of the Fab fragment of B2212A, a murine monoclonal antibody specific for the third fibronectin domain (Fn3) of human ROBO1. | | Descriptor: | anti-human ROBO1 antibody B2212A Fab heavy chain, anti-human ROBO1 antibody B2212A Fab light chain | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, S, Nakakido, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3B3R
 
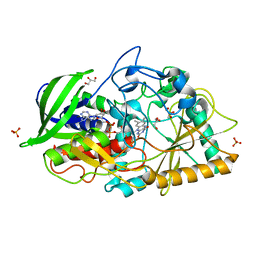 | | Crystal structure of Streptomyces cholesterol oxidase H447Q/E361Q mutant bound to glycerol (0.98A) | | Descriptor: | Cholesterol oxidase, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lyubimov, A.Y, Heard, K, Tang, H, Sampson, N.S, Vrielink, A. | | Deposit date: | 2007-10-22 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Distortion of flavin geometry is linked to ligand binding in cholesterol oxidase
Protein Sci., 16, 2007
|
|
3WIH
 
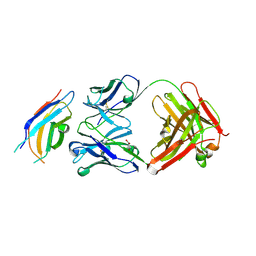 | | Crystal structure of the third fibronectin domain (Fn3) of human ROBO1 in complex with the Fab fragment of murine monoclonal antibody B2212A. | | Descriptor: | GLYCEROL, Roundabout homolog 1, anti-human ROBO1 antibody B2212A Fab heavy chain, ... | | Authors: | Nakayama, T, Mizohata, E, Yamashita, T, Nagatoishi, M, Iwanari, H, Mochizuki, Y, Kado, Y, Yokota, Y, Sato, R, Tsumoto, K, Fujitani, H, Kodama, T, Hamakubo, T, Inoue, T. | | Deposit date: | 2013-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural features of interfacial tyrosine residue in ROBO1 fibronectin domain-antibody complex: Crystallographic, thermodynamic, and molecular dynamic analyses
Protein Sci., 24, 2015
|
|
3WPD
 
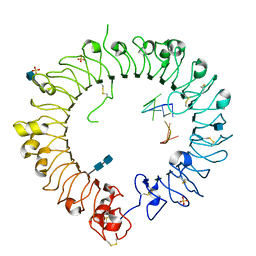 | | Crystal structure of horse TLR9 in complex with inhibitory DNA4084 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*CP*TP*GP*GP*AP*TP*GP*GP*G)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPC
 
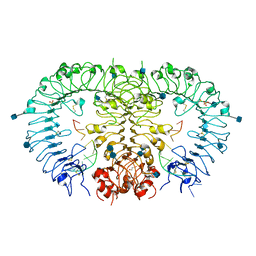 | | Crystal structure of horse TLR9 in complex with agonistic DNA1668_12mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*CP*AP*TP*GP*AP*CP*GP*TP*TP*CP*CP*T)-3'), ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPB
 
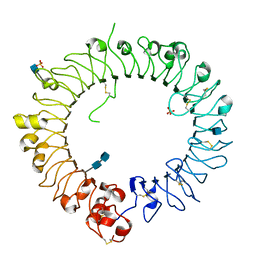 | | Crystal structure of horse TLR9 (unliganded form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Ohto, U, Tanji, H, Shimizu, T. | | Deposit date: | 2014-01-11 | | Release date: | 2015-02-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9
Nature, 520, 2015
|
|
3WPE
 
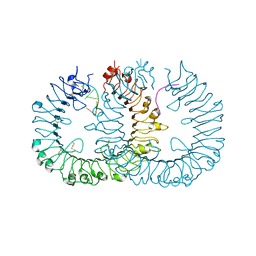 | |
7C4A
 
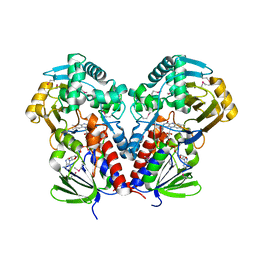 | | nicA2 with cofactor FAD | | Descriptor: | Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
7C49
 
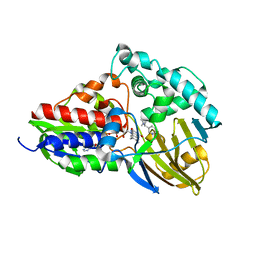 | | nicA2 with cofactor FAD and substrate nicotine | | Descriptor: | 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xu, P, Zhang, K. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Deceleration Regulates Toxicant Release to Prevent Cell Damage in Pseudomonas putida S16 (DSM 28022).
Mbio, 11, 2020
|
|
7EPM
 
 | | human LDHC complexed with NAD+ and ethylamino acetic acid | | Descriptor: | 2-(ethylamino)-2-oxidanylidene-ethanoic acid, L-lactate dehydrogenase C chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-04-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of human LDHC4 as a potential target for anticancer drug discovery.
Acta Pharm Sin B, 12, 2022
|
|
7U9S
 
 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | 5-{2-[4-(trifluoromethyl)phenyl]ethyl}-1,4-dihydropyrazine-2,3-dione, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
7U9U
 
 | | Crystal structure of human D-amino acid oxidase in complex with inhibitor | | Descriptor: | (3R)-3-(5,6-dioxo-1,4,5,6-tetrahydropyrazin-2-yl)-2,3-dihydro-1,4-benzoxathiine-7-carbonitrile, BENZOIC ACID, D-amino-acid oxidase, ... | | Authors: | Skene, R.J, Bell, J.A. | | Deposit date: | 2022-03-11 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of a Novel Class of d-Amino Acid Oxidase Inhibitors Using the Schrodinger Computational Platform.
J.Med.Chem., 65, 2022
|
|
5FLF
 
 | | DISEASE LINKED MUTATION IN FGFR | | Descriptor: | ACETATE ION, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1, ... | | Authors: | Thiyagarajan, N, Bunney, T.D, Katan, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Landscape of Activating Cancer Mutations in Fgfr Kinases and Their Differential Responses to Inhibitors in Clinical Use.
Oncotarget, 7, 2016
|
|
2CY1
 
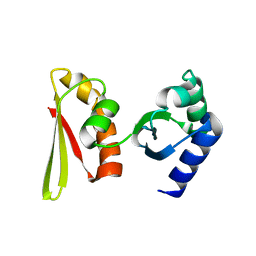 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3A07
 
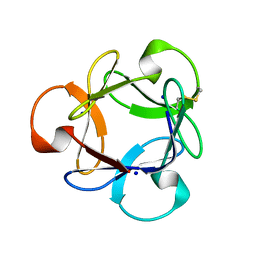 | | Crystal Structure of Actinohivin; Potent anti-HIV Protein | | Descriptor: | Actinohivin, SODIUM ION | | Authors: | Tsunoda, M, Suzuki, K, Sagara, T, Takenaka, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mechanism by which the lectin actinohivin blocks HIV infection of target cells
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5EW8
 
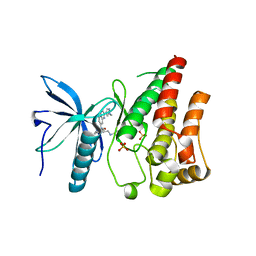 | | FIBROBLAST GROWTH FACTOR RECEPTOR 1 IN COMPLEX WITH JNJ-4275693 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}'-(3,5-dimethoxyphenyl)-~{N}'-[3-(1-methylpyrazol-4-yl)quinoxalin-6-yl]-~{N}-propan-2-yl-ethane-1,2-diamine | | Authors: | Ogg, D, Breed, J. | | Deposit date: | 2015-11-20 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Landscape of activating cancer mutations in FGFR kinases and their differential responses to inhibitors in clinical use.
Oncotarget, 7, 2016
|
|
7DPJ
 
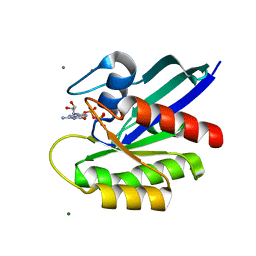 | | H-Ras Q61L in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Taniguchi, H, Matsumoto, S, Miyamoto, R, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7DPH
 
 | | H-Ras Q61H in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Taniguchi, H, Matsumoto, S, Kawamura, T, Kumasaka, T, Kataoka, T. | | Deposit date: | 2020-12-19 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Oncogenic mutations Q61L and Q61H confer active form-like structural features to the inactive state (state 1) conformation of H-Ras protein.
Biochem.Biophys.Res.Commun., 565, 2021
|
|
7E38
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with yatein and succinate | | Descriptor: | (3~{R},4~{R})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, (3~{S},4~{S})-4-(1,3-benzodioxol-5-ylmethyl)-3-[(3,4,5-trimethoxyphenyl)methyl]oxolan-2-one, Deoxypodophyllotoxin synthase, ... | | Authors: | Wu, M.-H, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7E37
 
 | | Crystal structure of deoxypodophyllotoxin synthase from Sinopodophyllum hexandrum in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Deoxypodophyllotoxin synthase, FE (III) ION | | Authors: | Wu, M.-H, Lin, H.-Y, Chang, W.-c, Chien, T.-C, Chan, N.-L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic analysis of carbon-carbon bond formation by deoxypodophyllotoxin synthase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
