6VV7
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB136 | | Descriptor: | 5-[3-(5-methyl-1H-indol-3-yl)propoxy]-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B, Chavez-Pacheco, S.M. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
2Y48
 
 | | Crystal structure of LSD1-CoREST in complex with a N-terminal SNAIL peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Baron, R, Binda, C, Tortorici, M, McCammon, J.A, Mattevi, A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Mimicry and Ligand Recognition in Binding and Catalysis by the Histone Demethylase Lsd1-Corest Complex.
Structure, 19, 2011
|
|
2XNI
 
 | | Protein-ligand complex of a novel macrocyclic HCV NS3 protease inhibitor derived from amino cyclic boronates | | Descriptor: | (1-{[(10-tert-butyl-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19,23,23a-tetradecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosin-7(3H)-yl)carbonyl]amino}-3-hydroxypropyl)(trihydroxy)borate(1-), MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Zhou, Y, Li, Q, Plattner, J.J, Baker, S.J, Zhang, S, Kazmierski, W.M, Wright, L.L, Smith, G.K, Grimes, R.M, Crosby, R.M, Creech, K.L, Carballo, L.H, Slater, M.J, Jarvest, R.L, Thommes, P, Hubbard, J.A, Convery, M.A, Nassau, P.M, McDowell, W, Skarzynski, T.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, Pennicott, L.E, Zou, W, Wright, J. | | Deposit date: | 2010-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Macrocyclic Hcv Ns3 Protease Inhibitors Derived from Alpha-Amino Cyclic Boronates.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XQC
 
 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | Descriptor: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
5A27
 
 | | Leishmania major N-myristoyltransferase in complex with a chlorophenyl 1,2,4-oxadiazole inhibitor. | | Descriptor: | 5-chloranyl-N-[2-(3-methoxyphenyl)ethanimidoyl]-2-piperidin-4-yloxy-benzamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Rackham, M.D, Yu, Z, Brannigan, J.A, Heal, W.P, Paape, D, Barker, K.V, Wilkinson, A.J, Smith, D.F, Tate, E.W, Leatherbarrow, R.J. | | Deposit date: | 2015-05-15 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery of High Affinity Inhibitors of Leishmania Donovani N-Myristoyltransferase.
Medchemcomm, 6, 2015
|
|
2XYY
 
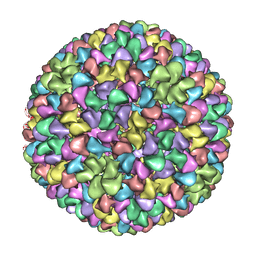 | | De Novo model of Bacteriophage P22 procapsid coat protein | | Descriptor: | COAT PROTEIN | | Authors: | Chen, D.-H, Baker, M.L, Hryc, C.F, DiMaio, F, Jakana, J, Wu, W, Dougherty, M, Haase-Pettingell, C, Schmid, M.F, Jiang, W, Baker, D, King, J.A, Chiu, W. | | Deposit date: | 2010-11-19 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Basis for Scaffolding-Mediated Assembly and Maturation of a DsDNA Virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6VV8
 
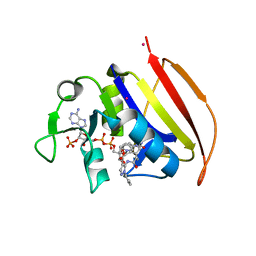 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with JEB285 | | Descriptor: | 5-{[3-(1H-indol-3-yl)propanoyl]amino}-1-phenyl-1H-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Chavez-Pacheco, S.M, Dias, M.V.B. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
5A46
 
 | | FGFR1 in complex with dovitinib | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Klein, T, Vajpai, N, Phillips, J.J, Davies, G, Holdgate, G.A, Phillips, C, Tucker, J.A, Norman, R.A, Scott, A.S, Higazi, D.R, Lowe, D, Thompson, G.S, Breeze, A.L. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Dynamic Insights Into the Energetics of Activation Loop Rearrangement in Fgfr1 Kinase.
Nat.Commun., 6, 2015
|
|
2Y2W
 
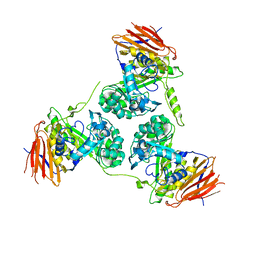 | | Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. | | Descriptor: | ARABINOFURANOSIDASE | | Authors: | Lagaert, S, Schoepe, J, Delcour, J.A, Lavigne, R, Strelkov, S.V, Courtin, C.M, Mikkelsen, N.E, Sandgren, M, Volckaert, G. | | Deposit date: | 2010-12-16 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elucidation of the Substrate Specificity and Protein Structure of Abfb, a Family 51 Alpha-L- Arabinofuranosidase from Bifidobacterium Longum.
To be Published
|
|
2XLJ
 
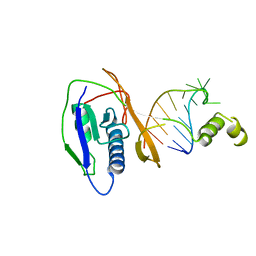 | | Crystal structure of the Csy4-crRNA complex, hexagonal form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-20 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
2XMA
 
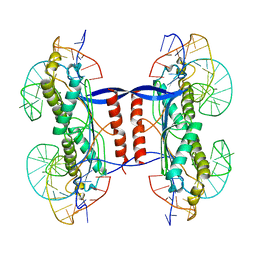 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
6U8K
 
 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV3 | | Descriptor: | Heavy chain of Fab HCV3, JIIIabc RNA (68-MER), Light chain of Fab HCV3 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
2XLK
 
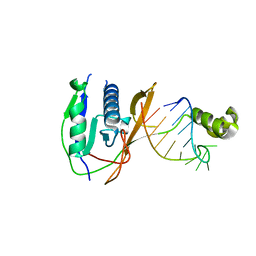 | | Crystal structure of the Csy4-crRNA complex, orthorhombic form | | Descriptor: | 5'-R(*CP*UP*GP*CP*CP*GP*UP*AP*UP*AP*GP*GP*CP*A*DG*C)-3', CSY4 ENDORIBONUCLEASE | | Authors: | Haurwitz, R.E, Jinek, M, Wiedenheft, B, Zhou, K, Doudna, J.A. | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Sequence- and Structure-Specific RNA Processing by a Crispr Endonuclease.
Science, 329, 2010
|
|
6VS5
 
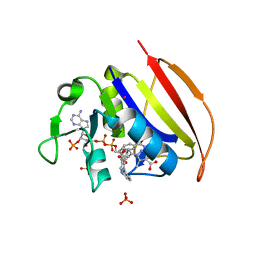 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 5-methyl-1-phenyl-1H-pyrazole-4-carboxylic acid (fragment 1) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Tyrakis, P, Blundell, T, Dias, M.V.B. | | Deposit date: | 2020-02-10 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
6VPC
 
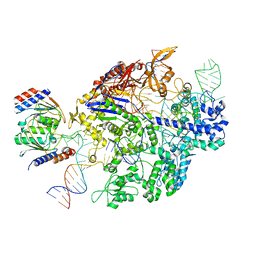 | | Structure of the SpCas9 DNA adenine base editor - ABE8e | | Descriptor: | CRISPR-associated endonuclease Cas9, Cas9 (SpCas9) single-guide RNA (sgRNA), DNA non-target strand (NTS), ... | | Authors: | Knott, G.J, Lapinaite, A, Doudna, J.A. | | Deposit date: | 2020-02-03 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | DNA capture by a CRISPR-Cas9-guided adenine base editor.
Science, 369, 2020
|
|
6VSE
 
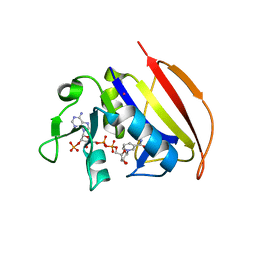 | |
5ANK
 
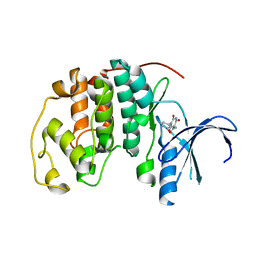 | | Crystal structure of CDK2 in complex with 2,4,6-trioxo-1-phenyl- hexahydropyrimidine-5-carboxamide processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | 2,4,6-TRIOXO-1-PHENYL-HEXAHYDROPYRIMIDINE-5-CARBOXAMIDE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
6UOQ
 
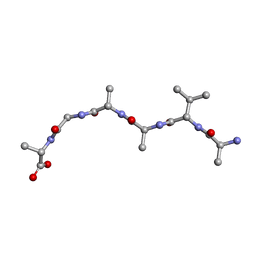 | |
5DFK
 
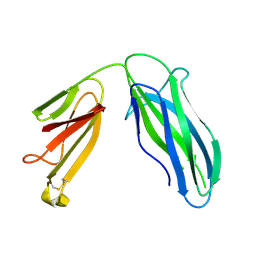 | |
2V78
 
 | | Crystal structure of Sulfolobus solfataricus 2-keto-3-deoxygluconate kinase | | Descriptor: | FRUCTOKINASE | | Authors: | Potter, J.A, Theodossis, A, Kerou, M, Lamble, H.J, Bull, S.D, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Sulfolobus Solfataricus 2-Keto-3-Deoxygluconate Kinase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5BRA
 
 | | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188 (Oant_2843, TARGET EFI-511085) | | Descriptor: | Putative periplasmic binding protein with substrate ribose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-30 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.971 Å) | | Cite: | Crystal Structure of a putative Periplasmic Solute binding protein (IPR025997) from Ochrobactrum Anthropi ATCC49188(Oant_2843, TARGET EFI-511085)
To be published
|
|
5BT3
 
 | | Crystal structure of EP300 bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase p300, ISOPROPYL ALCOHOL | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of EP300 bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
5BV5
 
 | | Structure of CYP119 with T213A and C317H mutations | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 119, PHOSPHATE ION, ... | | Authors: | Buller, A.R, Heel, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Adaptability Facilitates Histidine Heme Ligation in a Cytochrome P450.
J.Am.Chem.Soc., 137, 2015
|
|
6VSD
 
 | |
6VSG
 
 | | Mycobacterium tuberculosis dihydrofolate reductase in complex with 4-(trifluoromethyl)benzene-1,2-diamine(fragment 17) | | Descriptor: | 4-(TRIFLUOROMETHYL)BENZENE-1,2-DIAMINE, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Using a Fragment-Based Approach to Identify Alternative Chemical Scaffolds Targeting Dihydrofolate Reductase fromMycobacterium tuberculosis.
Acs Infect Dis., 6, 2020
|
|
