8HO2
 
 | |
2XCN
 
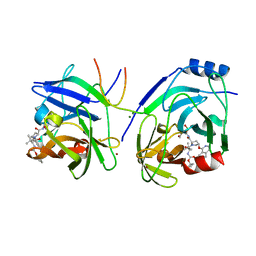 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | MAGNESIUM ION, N-[(CYCLOPENTYLOXY)CARBONYL]-3-METHYL-L-VALYL-(4R)-N-{(1R)-3-HYDROXY-1-[HYDROXY(OXIDO)BORANYL]PROPYL}-4-(ISOQUINOLIN-1-YLOXY)-L-PROLINAMIDE, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-23 | | Release date: | 2010-06-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCF
 
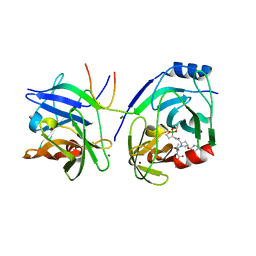 | | Crystal structure of HCV NS3 protease with a boronate inhibitor | | Descriptor: | CYCLOPENTYL N-[(2S)-1-[(2S,4R)-2-[[(4R)-8-HYDROXY-1,6,10-TRIOXA-5$L^{4}-BORASPIRO[4.5]DECAN-4-YL]CARBAMOYL]-4-ISOQUINOLIN-1-YLOXY-PYRROLIDIN-1-YL]-3,3-DIMETHYL-1-OXO-BUTAN-2-YL]CARBAMATE, MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Li, Q, Zhou, Y, Plattner, J.J, Baker, S.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, White, G.V, Mordaunt, J.E, Lazarides, L.X, Slater, M.J, Jarvest, R.L, Thommes, P, Ellis, M, Edge, C.M, Hubbard, J.A, Nassau, P, McDowell, B, Skarzynski, T.J, Rowland, P, Somers, D.O, Kazmierski, W.M, Grimes, R.M, Wright, L.L, Smith, G.K, Zou, W, Wright, J, Pennicott, L.E. | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-02 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis and Evaluation of Novel Alpha-Amino Cyclic Boronates as Inhibitors of Hcv Ns3 Protease.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XNI
 
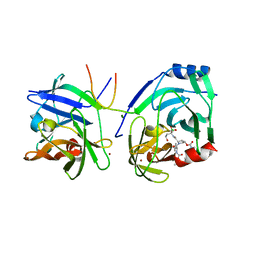 | | Protein-ligand complex of a novel macrocyclic HCV NS3 protease inhibitor derived from amino cyclic boronates | | Descriptor: | (1-{[(10-tert-butyl-15,15-dimethyl-3,9,12-trioxo-6,7,9,10,11,12,14,15,16,17,18,19,23,23a-tetradecahydro-1H,5H-2,23:5,8-dimethano-4,13,2,8,11-benzodioxatriazacyclohenicosin-7(3H)-yl)carbonyl]amino}-3-hydroxypropyl)(trihydroxy)borate(1-), MAGNESIUM ION, NS3 PROTEASE, ... | | Authors: | Li, X, Zhang, Y.-K, Liu, Y, Ding, C.Z, Zhou, Y, Li, Q, Plattner, J.J, Baker, S.J, Zhang, S, Kazmierski, W.M, Wright, L.L, Smith, G.K, Grimes, R.M, Crosby, R.M, Creech, K.L, Carballo, L.H, Slater, M.J, Jarvest, R.L, Thommes, P, Hubbard, J.A, Convery, M.A, Nassau, P.M, McDowell, W, Skarzynski, T.J, Qian, X, Fan, D, Liao, L, Ni, Z.-J, Pennicott, L.E, Zou, W, Wright, J. | | Deposit date: | 2010-08-02 | | Release date: | 2011-08-17 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Novel Macrocyclic Hcv Ns3 Protease Inhibitors Derived from Alpha-Amino Cyclic Boronates.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6Q08
 
 | |
2N0V
 
 | | Backbone 1H, Chemical Shift Assignments for Cn-APM1 | | Descriptor: | Antimicrobial peptide 1 | | Authors: | Santana, M.J, Oliveira, A.L, Queiroz Jr, L.K, Mandal, S.M, Matos, C.O, Dias, R.O, Franco, O.L, Liao, L.M. | | Deposit date: | 2015-03-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into Cn-AMP1, a short disulfide-free multifunctional peptide from green coconut water.
Febs Lett., 589, 2015
|
|
2N9R
 
 | | Novel antimicrobial peptide PaDBS1R1 designed from the ribosomal protein L39E from Pyrobaculum aerophilum using bioinformatics | | Descriptor: | Antimicrobial peptide PaDBS1R1 | | Authors: | Irazazabal, L.S.F, Porto, W.F, Alves, E.S.F, Matos, C.O, Hancock, R.E.W, Haney, E, Ribeiro, S.M, Liao, L.M, Ladram, A.S.F, Franco, O.L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-07 | | Method: | SOLUTION NMR | | Cite: | Novel antimicrobial peptide PaDBS1R1 designed from the ribosomal protein L39E from Pyrobaculum aerophilum using bioinformatics
To be Published
|
|
8ERU
 
 | | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R4 | | Descriptor: | Mastoparan-R4 peptide | | Authors: | Freitas, C.D.P, Oshiro, K.G.N, Macedo, M.L.R, Cardoso, M.H, Franco, O.L, Liao, L.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a computacionally designed mastoparan-like peptide, mastoparan-R4
To Be Published
|
|
8EP5
 
 | | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R1 | | Descriptor: | Mastoparan-R1 peptide | | Authors: | Freitas, C.D.P, Oshiro, K.G.N, Macedo, M.L.R, Cardoso, M.H, Franco, O.L, Liao, L.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-18 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R1.
To Be Published
|
|
6DUL
 
 | |
6DUU
 
 | |
6D2H
 
 | |
7T7W
 
 | |
8IWK
 
 | | ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ABCG25 Wild Type purified with DDM plus CHS in ABA-bound state
To Be Published
|
|
8IWN
 
 | | ABCG25 EQ mutant in ATP-bound state | | Descriptor: | ABC transporter G family member 25, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, L, Liu, X, Ying, W, Liao, L, Wei, H. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | ABCG25 EQ mutant in ATP-bound state
To Be Published
|
|
8IWJ
 
 | |
7DAL
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAJ
 
 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6CFA
 
 | | peptide PaAMP1R3 | | Descriptor: | peptide PaAMP1R3 | | Authors: | Alves, E.S.F, Liao, L.M. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Synthetic peptide PaAMP1R3
To be Published
|
|
5J6T
 
 | |
5J6W
 
 | |
5J6V
 
 | |
7Y6C
 
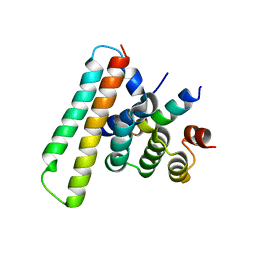 | | Crystal structure of the EscE/EsaG/EsaH complex | | Descriptor: | EscE/YscE/SsaE family type III secretion system needle protein co-chaperone, EscG/YscG/SsaH family type III secretion system needle protein co-chaperone | | Authors: | Zeng, Z.X, Xiao, S.B, Liao, L.J, Zhou, Y.Z. | | Deposit date: | 2022-06-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Secreted in a Type III Secretion System-Dependent Manner, EsaH and EscE Are the Cochaperones of the T3SS Needle Protein EsaG of Edwardsiella piscicida.
Mbio, 13, 2022
|
|
