2V64
 
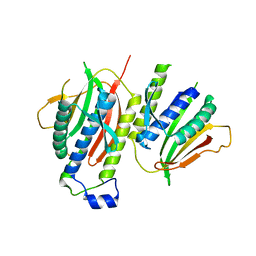 | | Crystallographic structure of the conformational dimer of the Spindle Assembly Checkpoint protein Mad2. | | 分子名称: | MBP1, MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A | | 著者 | Mapelli, M, Massimiliano, L, Santaguida, S, Musacchio, A. | | 登録日 | 2007-07-13 | | 公開日 | 2007-11-27 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The MAD2 Conformational Dimer: Structure and Implications for the Spindle Assembly Checkpoint
Cell(Cambridge,Mass.), 131, 2007
|
|
1UNL
 
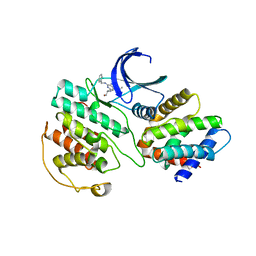 | | Structural mechanism for the inhibition of CD5-p25 from the roscovitine, aloisine and indirubin. | | 分子名称: | CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1, R-ROSCOVITINE | | 著者 | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | 登録日 | 2003-09-10 | | 公開日 | 2004-11-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNG
 
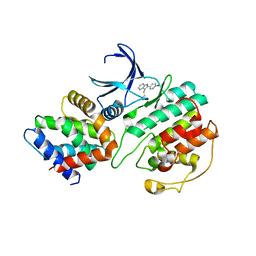 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | 分子名称: | 6-PHENYL[5H]PYRROLO[2,3-B]PYRAZINE, CELL DIVISION PROTEIN KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | 著者 | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | 登録日 | 2003-09-10 | | 公開日 | 2004-11-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
1UNH
 
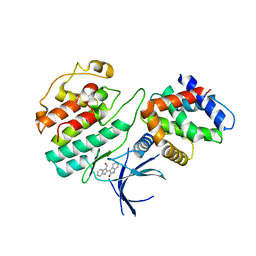 | | Structural mechanism for the inhibition of CDK5-p25 by roscovitine, aloisine and indirubin. | | 分子名称: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, CYCLIN-DEPENDENT KINASE 5, CYCLIN-DEPENDENT KINASE 5 ACTIVATOR 1 | | 著者 | Mapelli, M, Crovace, C, Massimiliano, L, Musacchio, A. | | 登録日 | 2003-09-10 | | 公開日 | 2004-11-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Mechanism of Cdk5/P25 Binding by Cdk Inhibitors
J.Med.Chem., 48, 2005
|
|
3O0G
 
 | | Crystal Structure of Cdk5:p25 in complex with an ATP analogue | | 分子名称: | Cell division protein kinase 5, Cyclin-dependent kinase 5 activator 1, {4-amino-2-[(4-chlorophenyl)amino]-1,3-thiazol-5-yl}(3-nitrophenyl)methanone | | 著者 | Mapelli, M. | | 登録日 | 2010-07-19 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Defining Cdk5 ligand chemical space with small molecule inhibitors of Tau phosphorylation
Chem.Biol., 12, 2005
|
|
5NJJ
 
 | | PTB domain of human Numb isoform-1 | | 分子名称: | ALA-TYR-ILE-GLY-PRO-PTR-LEU, Protein numb homolog, SULFATE ION | | 著者 | Mapelli, M, Di Fiore, P.P. | | 登録日 | 2017-03-29 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A Numb-Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer.
J. Cell Biol., 217, 2018
|
|
5NJK
 
 | | PTB domain of human Numb isoform-1 | | 分子名称: | ALA-TYR-ILE-GLY-PRO-PTR-LEU, Protein numb homolog, SULFATE ION | | 著者 | Mapelli, M, Di Fiore, P.P. | | 登録日 | 2017-03-29 | | 公開日 | 2017-12-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | A Numb-Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer.
J. Cell Biol., 217, 2018
|
|
1URJ
 
 | | Single stranded DNA-binding protein(ICP8) from Herpes simplex virus-1 | | 分子名称: | MAJOR DNA-BINDING PROTEIN, MERCURY (II) ION, ZINC ION | | 著者 | Panjikar, S, Mapelli, M, Tucker, P.A. | | 登録日 | 2003-10-30 | | 公開日 | 2004-11-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the herpes simplex virus 1 ssDNA-binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding.
J. Biol. Chem., 280, 2005
|
|
6QJA
 
 | | Organizational principles of the NuMA-Dynein interaction interface and implications for mitotic spindle functions | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Nuclear mitotic apparatus protein 1 | | 著者 | Renna, C, Rizzelli, F, Carminati, M, Gaddoni, C, Pirovano, L, Cecatiello, V, Pasqualato, S, Mapelli, M. | | 登録日 | 2019-01-23 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Organizational Principles of the NuMA-Dynein Interaction Interface and Implications for Mitotic Spindle Functions.
Structure, 28, 2020
|
|
6HC2
 
 | | Crystal structure of NuMA/LGN hetero-hexamers | | 分子名称: | G-protein-signaling modulator 2, Nuclear mitotic apparatus protein 1 | | 著者 | Pasqualato, S, Culurgioni, S, Foadi, J, Alfieri, A, Mapelli, M. | | 登録日 | 2018-08-13 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (4.31 Å) | | 主引用文献 | Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions.
Nat Commun, 10, 2019
|
|
5A7D
 
 | | Tetrameric assembly of LGN with Inscuteable | | 分子名称: | INSCUTEABLE, PINS | | 著者 | Culurgioni, S, Mari, S, Bonetto, G, Gallini, S, Brennich, M, Round, A, Mapelli, M. | | 登録日 | 2015-07-07 | | 公開日 | 2016-08-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | The Structure of the Insc:Lgn Tetramer Reveals a New Function of Lgn in Promoting Asymmetric Cell Divisions
To be Published
|
|
5A6C
 
 | | Concomitant binding of Afadin to LGN and F-actin directs planar spindle orientation | | 分子名称: | G-PROTEIN-SIGNALING MODULATOR 2, AFADIN, SULFATE ION | | 著者 | Carminati, M, Gallini, S, Pirovano, L, Alfieri, A, Bisi, S, Mapelli, M. | | 登録日 | 2015-06-25 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | Concomitant Binding of Afadin to Lgn and F-Actin Directs Planar Spindle Orientation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4A7J
 
 | | Symmetric Dimethylation of H3 Arginine 2 is a Novel Histone Mark that Supports Euchromatin Maintenance | | 分子名称: | HISTONE H3.1T, WD REPEAT-CONTAINING PROTEIN 5 | | 著者 | Migliori, V, Muller, J, Phalke, S, Low, D, Bezzi, M, ChuenMok, W, Gunaratne, J, Capasso, P, Bassi, C, Cecatiello, V, DeMarco, A, Blackstock, W, Kuznetsov, V, Amati, B, Mapelli, M, Guccione, E. | | 登録日 | 2011-11-14 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Symmetric Dimethylation of H3R2 is a Newly Identified Histone Mark that Supports Euchromatin Maintenance
Nat.Struct.Mol.Biol., 19, 2012
|
|
4A1S
 
 | | Crystallographic structure of the Pins:Insc complex | | 分子名称: | PARTNER OF INSCUTEABLE, RE60102P | | 著者 | Culurgioni, S, Alfieri, A, Pendolino, V, Laddomada, F, Mapelli, M. | | 登録日 | 2011-09-19 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inscuteable and Numa Proteins Bind Competitively to Leu-Gly- Asn Repeat-Enriched Protein (Lgn) During Asymmetric Cell Divisions.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2C7M
 
 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | 分子名称: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | 著者 | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | 登録日 | 2005-11-25 | | 公開日 | 2006-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin.
Cell, 124, 2006
|
|
6ZT0
 
 | | Crystal structure of the Eiger TNF domain/Grindelwald extracellular domain complex | | 分子名称: | 1,2-ETHANEDIOL, Protein eiger, Protein grindelwald | | 著者 | Palmerini, V, Cecatiello, V, Pasqualato, S, Mapelli, M. | | 登録日 | 2020-07-17 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions.
Nat Commun, 12, 2021
|
|
6ZSZ
 
 | |
6ZSY
 
 | |
1GO4
 
 | | Crystal structure of Mad1-Mad2 reveals a conserved Mad2 binding motif in Mad1 and Cdc20. | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD1, Mitotic spindle assembly checkpoint protein MAD2A | | 著者 | Sironi, L, Mapelli, M, Jeang, K.T, Musacchio, A. | | 登録日 | 2001-10-17 | | 公開日 | 2002-05-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint.
EMBO J., 21, 2002
|
|
2BFY
 
 | | Complex of Aurora-B with INCENP and Hesperadin. | | 分子名称: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A, N-[2-OXO-3-((E)-PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]IMINO}METHYL)-2,6-DIHYDRO-1H-INDOL-5-YL]ETHANESULFONAMIDE | | 著者 | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | 登録日 | 2004-12-15 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
2BFX
 
 | | Mechanism of Aurora-B activation by INCENP and inhibition by Hesperadin. | | 分子名称: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A | | 著者 | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | 登録日 | 2004-12-15 | | 公開日 | 2005-05-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
2C7N
 
 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | 分子名称: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | 著者 | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | 登録日 | 2005-11-25 | | 公開日 | 2006-02-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Ubiquitin Binding Domains of Rabex-5 Reveals Two Modes of Interaction with Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2N12
 
 | |
