8EDT
 
 | |
8EDS
 
 | |
8EDR
 
 | |
4YNG
 
 | | Twinned pyruvate kinase from E. coli in the T-state | | 分子名称: | Pyruvate kinase I, SULFATE ION | | 著者 | Donovan, K.A, Dobson, R.C.J. | | 登録日 | 2015-03-10 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Grappling with anisotropic data, pseudo-merohedral twinning and pseudo-translational noncrystallographic symmetry: a case study involving pyruvate kinase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8EQ0
 
 | |
8EQ1
 
 | |
8EQ3
 
 | | Escherichia coli pyruvate kinase A301T | | 分子名称: | IMIDAZOLE, MALONATE ION, Pyruvate kinase, ... | | 著者 | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | 登録日 | 2022-10-07 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
8EU4
 
 | |
8EDQ
 
 | | E. coli pyruvate kinase (PykF) I264F | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Pyruvate kinase, SULFATE ION | | 著者 | Donovan, K.A, Coombes, D, Dobson, R.C.J, Cooper, T.F. | | 登録日 | 2022-09-05 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Beneficial mutations occurring in E. coli pyruvate kinase afford new allosteric mechanisms leading to faster resumption of growth
To Be Published
|
|
6Q0W
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0R
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to E7820 and RBM39 | | 分子名称: | 3-cyano-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
6Q0V
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to tasisulam and RBM39 | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | 著者 | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | 登録日 | 2019-08-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
7MEU
 
 | |
7RCT
 
 | |
7LPS
 
 | | Crystal structure of DDB1-CRBN-ALV1 complex bound to Helios (IKZF2 ZF2) | | 分子名称: | 3-[3-[[1-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(oxidanylidene)pyrrol-3-yl]amino]phenyl]-~{N}-(3-chloranyl-4-methyl-phenyl)propanamide, DNA damage-binding protein 1, Protein cereblon, ... | | 著者 | Nowak, R.P, Fischer, E.S. | | 登録日 | 2021-02-12 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.78 Å) | | 主引用文献 | Acute pharmacological degradation of Helios destabilizes regulatory T cells.
Nat.Chem.Biol., 17, 2021
|
|
6TD3
 
 | | Structure of DDB1 bound to CR8-engaged CDK12-cyclinK | | 分子名称: | (2R)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Bunker, R.D, Petzold, G, Kozicka, Z, Thoma, N.H. | | 登録日 | 2019-11-07 | | 公開日 | 2020-06-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.46 Å) | | 主引用文献 | The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Nature, 585, 2020
|
|
8P82
 
 | | Cryo-EM structure of dimeric UBR5 | | 分子名称: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | 著者 | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-05-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
8P83
 
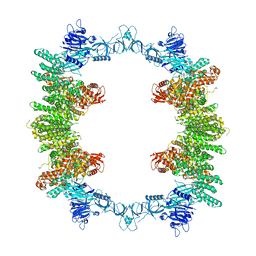 | | Cryo-EM structure of full-length human UBR5 (homotetramer) | | 分子名称: | E3 ubiquitin-protein ligase UBR5 | | 著者 | Aguirre, J.D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | 登録日 | 2023-05-31 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | UBR5 forms ligand-dependent complexes on chromatin to regulate nuclear hormone receptor stability.
Mol.Cell, 83, 2023
|
|
6N2F
 
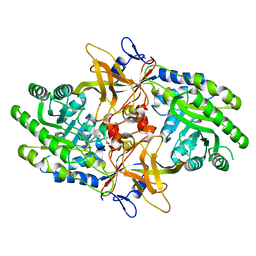 | |
6N2A
 
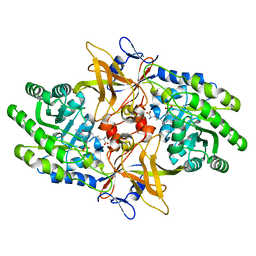 | |
8G46
 
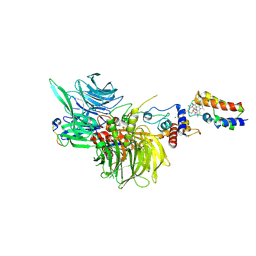 | | Cryo-EM structure of DDB1deltaB-DDA1-DCAF16-BRD4(BD2)-MMH2 | | 分子名称: | Bromodomain-containing protein 4, DDB1- and CUL4-associated factor 16, DET1- and DDB1-associated protein 1, ... | | 著者 | Ma, M.W, Hunkeler, M, Jin, C.Y, Fischer, E.S. | | 登録日 | 2023-02-08 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Template-assisted covalent modification of DCAF16 underlies activity of BRD4 molecular glue degraders.
Biorxiv, 2023
|
|
6XMX
 
 | | Cryo-EM structure of BCL6 bound to BI-3802 | | 分子名称: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | 著者 | Yoon, H, Burman, S.S.R, Hunkeler, M, Nowak, R.P, Fischer, E.S. | | 登録日 | 2020-07-01 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Small-molecule-induced polymerization triggers degradation of BCL6.
Nature, 588, 2020
|
|
8BUF
 
 | | Structure of DDB1 bound to Z12-engaged CDK12-cyclin K | | 分子名称: | 2-(6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-ylmethyl)-6,7-dimethoxy-3~{H}-quinazolin-4-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUJ
 
 | | Structure of DDB1 bound to DS06-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-(octylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (3.62 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU1
 
 | | Structure of DDB1 bound to DS17-engaged CDK12-cyclin K | | 分子名称: | (2~{R})-2-[[6-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methylamino]-9-(1-methylpyrazol-4-yl)purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | 著者 | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | 登録日 | 2022-11-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
