2P5E
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC Reveal Native Diagonal Binding Geometry | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-03-15 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | 著者 | Zhao, Z, Qi, J, Gao, F.G. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
2P0C
 
 | | Catalytic Domain of the Proto-oncogene Tyrosine-protein Kinase MER | | 分子名称: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Walker, J.R, Huang, X, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-02-28 | | 公開日 | 2007-05-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the inhibited states of the Mer receptor tyrosine kinase.
J.Struct.Biol., 165, 2009
|
|
4E4K
 
 | | Crystal Structure of PPARgamma with the ligand JO21 | | 分子名称: | (2S)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | 登録日 | 2012-03-13 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4E4Q
 
 | | Crystal structure of PPARgamma with the ligand FS214 | | 分子名称: | (2R)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | 登録日 | 2012-03-13 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | 著者 | Liu, C, Song, D, Dou, C. | | 登録日 | 2021-02-04 | | 公開日 | 2021-05-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
7EMK
 
 | | Dendrorhynchus zhejiangensis ferritin | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dendrorhynchus zhejiangensis ferritin, ... | | 著者 | Huan, H.S, Ming, T.H, Su, X.R. | | 登録日 | 2021-04-14 | | 公開日 | 2021-05-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure determination of ferritin from Dendrorhynchus zhejiangensis
Biochemical and Biophysical Research Communications, 531, 2020
|
|
6K9H
 
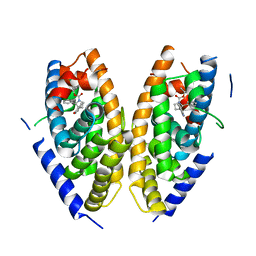 | | Human LXR-beta in complex with an agonist | | 分子名称: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | 著者 | Zhang, Z, Zhou, H. | | 登録日 | 2019-06-15 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
1UU0
 
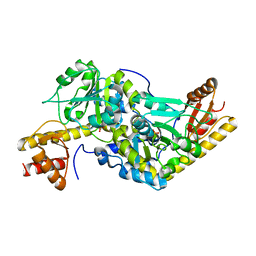 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (Apo-form) | | 分子名称: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHATE ION | | 著者 | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | 登録日 | 2003-12-12 | | 公開日 | 2004-05-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
1UU2
 
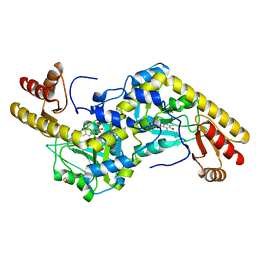 | | Histidinol-phosphate aminotransferase (HisC) from Thermotoga maritima (apo-form) | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | 著者 | Vega, M.C, Fernandez, F.J, Lehmann, F, Wilmanns, M. | | 登録日 | 2003-12-13 | | 公開日 | 2004-03-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural Studies of the Catalytic Reaction Pathway of a Hyperthermophilic Histidinol-Phosphate Aminotransferase
J.Biol.Chem., 279, 2004
|
|
2PYF
 
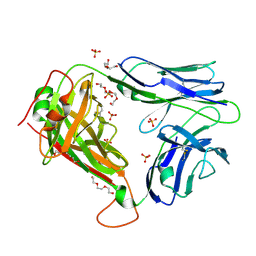 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry Unbound TCR Clone 5-1 | | 分子名称: | SULFATE ION, T-Cell Receptor, Alpha Chain, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-05-16 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
1CJL
 
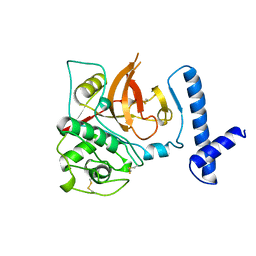 | |
1R8K
 
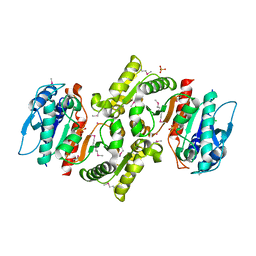 | | PDXA PROTEIN; NAD-DEPENDENT DEHYDROGENASE/CARBOXYLASE; SUBUNIT OF PYRIDOXINE PHOSPHATE BIOSYNTHETIC PROTEIN PDXJ-PDXA [SALMONELLA TYPHIMURIUM] | | 分子名称: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, CHLORIDE ION, COBALT (II) ION, ... | | 著者 | Osipiuk, J, Quartey, P, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-10-27 | | 公開日 | 2003-11-11 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of NAD-dependent dehydrogenase/carboxylase of Salmonella typhimurium
to be published
|
|
1DIB
 
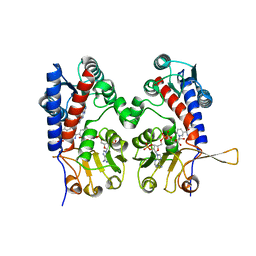 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | 分子名称: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | 登録日 | 1999-11-29 | | 公開日 | 2000-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIG
 
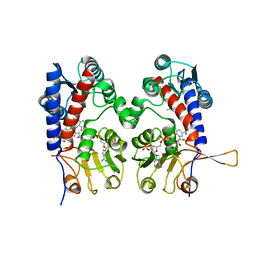 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY374571 | | 分子名称: | ACETATE ION, METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | 登録日 | 1999-11-29 | | 公開日 | 2000-07-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
6K9G
 
 | | Human LXR-beta in complex with an agonist | | 分子名称: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{R},3~{R})-2'-[3-[4-(hydroxymethyl)-3-methylsulfonyl-phenyl]phenyl]-2-oxidanylidene-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | 著者 | Zhang, Z, Zhou, H. | | 登録日 | 2019-06-15 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
2PYE
 
 | | Crystal Structures of High Affinity Human T-Cell Receptors Bound to pMHC RevealNative Diagonal Binding Geometry TCR Clone C5C1 Complexed with MHC | | 分子名称: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Beta-2-microglobulin, Cancer/testis antigen 1B, ... | | 著者 | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | 登録日 | 2007-05-16 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
