6LJW
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-phenylazanylbenzoic acid, Fatty acid-binding protein, ... | | 著者 | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | 登録日 | 2019-12-17 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
2H2G
 
 | |
7LC3
 
 | | CryoEM Structure of KdpFABC in E1-ATP state | | 分子名称: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | 登録日 | 2021-01-09 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2H2I
 
 | | The Structural basis of Sirtuin Substrate Affinity | | 分子名称: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, NAD-dependent deacetylase, ZINC ION | | 著者 | Cosgrove, M.S, Wolberger, C. | | 登録日 | 2006-05-18 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of sirtuin substrate affinity
Biochemistry, 45, 2006
|
|
7LC6
 
 | | Cryo-EM Structure of KdpFABC in E2-P state with BeF3 | | 分子名称: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | 登録日 | 2021-01-09 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2H2H
 
 | |
6KKH
 
 | | Crystal structure of the oxalate bound malyl-CoA lyase from Roseiflexus castenholzii | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | 著者 | Tang, W.R, Wang, Z.G, Zhang, C.Y, Wang, C. | | 登録日 | 2019-07-25 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
2FDW
 
 | | Crystal Structure Of Human Microsomal P450 2A6 with the inhibitor (5-(Pyridin-3-yl)furan-2-yl)methanamine bound | | 分子名称: | (5-(PYRIDIN-3-YL)FURAN-2-YL)METHANAMINE, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yano, J.K, Stout, C.D, Johnson, E.F. | | 登録日 | 2005-12-14 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Synthetic Inhibitors of Cytochrome P-450 2A6: Inhibitory Activity, Difference Spectra, Mechanism of Inhibition, and Protein Cocrystallization.
J.Med.Chem., 49, 2006
|
|
6JNY
 
 | |
8HWT
 
 | |
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | 著者 | He, Q.W, Xu, Z.P, Xie, Y.F. | | 登録日 | 2023-01-02 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
8HZ5
 
 | |
8HZ4
 
 | |
6UEG
 
 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | 分子名称: | 3-({2-[(2R)-2-carbamoyl-2,3-dihydro-4H-1,4-benzoxazin-4-yl]-2-oxoethyl}sulfanyl)propanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CALCIUM ION | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
5WYH
 
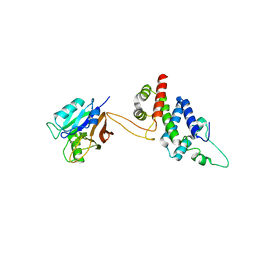 | |
6UEC
 
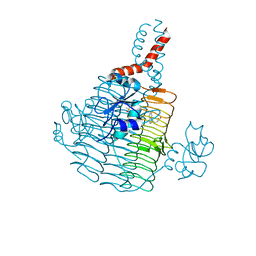 | | Pseudomonas aeruginosa LpxD Complex Structure with Ligand | | 分子名称: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UEE
 
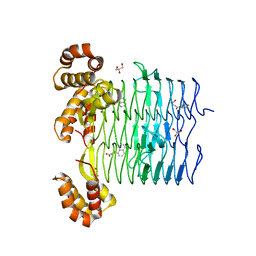 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | 分子名称: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, GLYCEROL | | 著者 | Chen, Y, Kroeck, K, Sacco, M. | | 登録日 | 2019-09-20 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | 分子名称: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
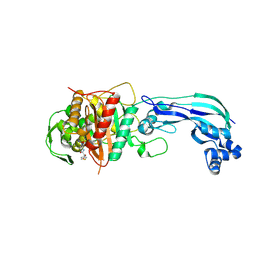 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | 分子名称: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Sacco, M, Chen, Y. | | 登録日 | 2019-10-10 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
4RF3
 
 | | Crystal Structure of ketoreductase from Lactobacillus kefir, mutant A94F | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | 著者 | Tang, Y, Tibrewal, N, Cascio, D. | | 登録日 | 2014-09-24 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.694 Å) | | 主引用文献 | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7WJQ
 
 | |
4RF5
 
 | | Crystal structure of ketoreductase from Lactobacillus kefir, E145S mutant | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | 著者 | Tang, Y, Tibrewal, N, Cascio, D. | | 登録日 | 2014-09-24 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8DWB
 
 | |
8DMK
 
 | |
4RF2
 
 | |
