5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | 分子名称: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | 著者 | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | 登録日 | 2015-08-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
9B4E
 
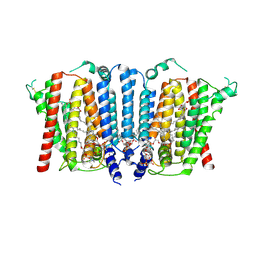 | | Structure of wild type human PSS1 | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CALCIUM ION, ... | | 著者 | Long, T, Li, X. | | 登録日 | 2024-03-20 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 2024
|
|
9B4G
 
 | | Structure of inhibitor-bound human PSS1 | | 分子名称: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, (7P)-7-[(4S)-4-{4-[3,5-bis(trifluoromethyl)phenoxy]phenyl}-5-(2,2-difluoropropyl)-6-oxo-1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazol-3-yl]-1,3-benzoxazol-2(3H)-one, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | 著者 | Long, T, Li, X. | | 登録日 | 2024-03-20 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 2024
|
|
9B4F
 
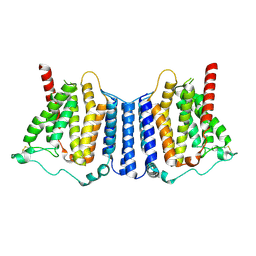 | | Structure of human PSS1-P269S | | 分子名称: | CALCIUM ION, Phosphatidylserine synthase 1 | | 著者 | Long, T, Li, X. | | 登録日 | 2024-03-20 | | 公開日 | 2024-09-04 | | 最終更新日 | 2024-09-11 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Molecular insights into human phosphatidylserine synthase 1 reveal its inhibition promotes LDL uptake.
Cell, 2024
|
|
7RR5
 
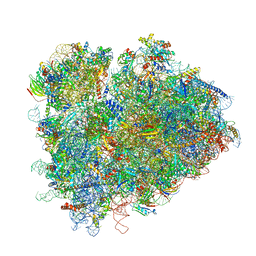 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | 著者 | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | 登録日 | 2021-08-09 | | 公開日 | 2021-11-10 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
3ZIF
 
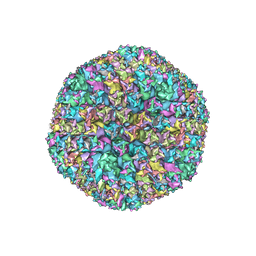 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | 分子名称: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | 著者 | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | 登録日 | 2013-01-09 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
6E1H
 
 | | Structure of 2:1 human Ptch1-Shh-N complex | | 分子名称: | CALCIUM ION, PALMITIC ACID, Protein patched homolog 1, ... | | 著者 | Qi, X, Li, X. | | 登録日 | 2018-07-09 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex.
Science, 362, 2018
|
|
8YC0
 
 | | T cell receptor V delta2 V gamma9 in GDN | | 分子名称: | CHOLESTEROL, T cell receptor delta variable 2,T cell receptor delta constant, T cell receptor gamma variable 9,T cell receptor gamma constant 1, ... | | 著者 | Xin, W, Huang, B, Chi, X, Xu, M, Zhang, Y, Li, X, Su, Q, Zhou, Q. | | 登録日 | 2024-02-17 | | 公開日 | 2024-05-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Structures of human gamma delta T cell receptor-CD3 complex.
Nature, 630, 2024
|
|
8PNV
 
 | |
8PNU
 
 | |
8Y6V
 
 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | 分子名称: | gp119, gp120, gp162, ... | | 著者 | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | 登録日 | 2024-02-03 | | 公開日 | 2024-08-14 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
5TJB
 
 | | I-II linker of TRPML1 channel at pH 4.5 | | 分子名称: | Mucolipin-1 | | 著者 | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | 登録日 | 2016-10-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3QQ3
 
 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | 分子名称: | 9-mer peptide from Neuraminidase, Beta-2-microglobulin, MHC class I antigen | | 著者 | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | 登録日 | 2011-02-15 | | 公開日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
5TJA
 
 | | I-II linker of TRPML1 channel at pH 6 | | 分子名称: | Mucolipin-1 | | 著者 | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | 登録日 | 2016-10-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5TJC
 
 | | I-II linker of TRPML1 channel at pH 7.5 | | 分子名称: | Mucolipin-1 | | 著者 | Li, M, Zhang, W.K, Benvin, N.M, Zhou, X, Su, D, Li, H, Wang, S, Michailidis, I.E, Tong, L, Li, X, Yang, J. | | 登録日 | 2016-10-04 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3CTM
 
 | | Crystal Structure of a Carbonyl Reductase from Candida Parapsilosis with anti-Prelog Stereo-specificity | | 分子名称: | Carbonyl Reductase | | 著者 | Zhang, R, Zhu, G, Li, X, Xu, Y, Zhang, X.C, Rao, Z. | | 登録日 | 2008-04-14 | | 公開日 | 2008-05-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Crystal structure of a carbonyl reductase from Candida parapsilosis with anti-Prelog stereospecificity.
Protein Sci., 17, 2008
|
|
3QQ4
 
 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, VP35 | | 著者 | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | 登録日 | 2011-02-15 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
4Q2V
 
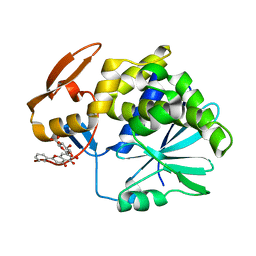 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | 分子名称: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | 著者 | Deng, X, Li, X, Dong, J, Chen, Y. | | 登録日 | 2014-04-10 | | 公開日 | 2015-04-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
8Z4L
 
 | |
8Z9E
 
 | |
8Z9C
 
 | |
8Z99
 
 | |
8YHE
 
 | |
8YHD
 
 | |
8Z4J
 
 | |
