4LCH
 
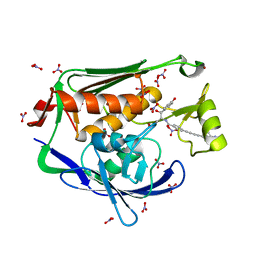 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-051 complex | | 分子名称: | (betaS)-Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N,beta-dihydroxy-beta-methyl-L-tyrosinamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
4LCF
 
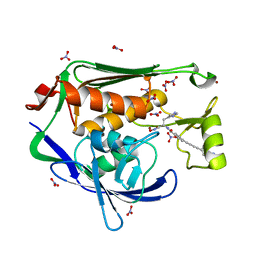 | | Crystal structure of the Pseudomonas aeruginosa LPXC/LPC-014 complex | | 分子名称: | NITRATE ION, Nalpha-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-histidinamide, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | 著者 | Lee, C.-J, Zhou, P. | | 登録日 | 2013-06-21 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Synthesis, Structure, and Antibiotic Activity of Aryl-Substituted LpxC Inhibitors.
J.Med.Chem., 56, 2013
|
|
8IVQ
 
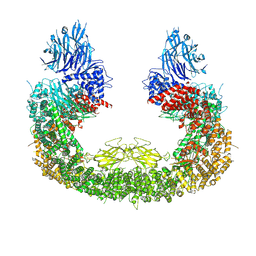 | | Cryo-EM structure of mouse BIRC6, Global map | | 分子名称: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | 著者 | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | 登録日 | 2023-03-28 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
4LI4
 
 | |
4LM9
 
 | |
4LMC
 
 | |
4LM7
 
 | |
9MI7
 
 | | Crystal Structure of ADI-64597 ((human Fab, with substituted IgG1-CH1 (HC-L128R and K147R) and substituted kappa constant domain (LC-Q124E, V133Q, and T178R)) | | 分子名称: | ADI-64597 Fab heavy chain, ADI-64597 Fab light chain, CHLORIDE ION, ... | | 著者 | Battles, M.B, Welin, M, Laursen, M. | | 登録日 | 2024-12-12 | | 公開日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Design of orthogonal constant domain interfaces to aid proper heavy/light chain pairing of bispecific antibodies.
Mabs, 17, 2025
|
|
9MFN
 
 | | Crystal Structure of ADI-64596 (human Fab, with substituted IgG1-CH1 (HC-L145Q, K147E, and S181E) and substituted kappa constant domain (LC-T129R, T178R, and T180Q)) | | 分子名称: | ADI-64596 Fab heavy chain, ADI-64596 Fab light chain, BROMIDE ION, ... | | 著者 | Battles, M.B, Welin, M, Laursen, M. | | 登録日 | 2024-12-10 | | 公開日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Design of orthogonal constant domain interfaces to aid proper heavy/light chain pairing of bispecific antibodies.
Mabs, 17, 2025
|
|
8Q91
 
 | | Structure of the human 20S U5 snRNP core | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Schneider, S, Galej, W.P. | | 登録日 | 2023-08-19 | | 公開日 | 2024-03-27 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6UD7
 
 | | Crystal structure of full-length human DCAF15-DDB1(deltaBPB)-DDA1-RBM39 in complex with indisulam | | 分子名称: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1,DNA damage-binding protein 1, ... | | 著者 | Bussiere, D.E, Shu, W, Xie, L, Knapp, M. | | 登録日 | 2019-09-18 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6UE5
 
 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | 分子名称: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | 著者 | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | 登録日 | 2019-09-20 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6WJC
 
 | |
2F5J
 
 | |
3CWJ
 
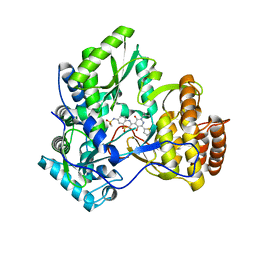 | | Crystal structure of hcv ns5b polymerase with a novel pyridazinone inhibitor | | 分子名称: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | 著者 | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | 登録日 | 2008-04-21 | | 公開日 | 2009-04-21 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 4-(1,1-Dioxo-1,4-dihydro-1lambda6-benzo[1,4]thiazin-3-yl)-5-hydroxy-2H-pyridazin-3-ones as potent inhibitors of HCV NS5B polymerase
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6WC5
 
 | |
6WC2
 
 | |
7FCP
 
 | |
7FCQ
 
 | |
5B7P
 
 | |
5B7G
 
 | |
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | 分子名称: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | 著者 | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | 登録日 | 2015-07-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
5B7N
 
 | |
5B7Q
 
 | |
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | 著者 | Zhu, J, Xu, T, Feng, B, Liu, J. | | 登録日 | 2021-07-11 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
