1LBV
 
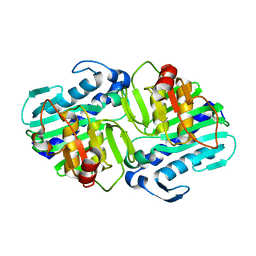 | | Crystal Structure of apo-form (P21) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | 分子名称: | fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBW
 
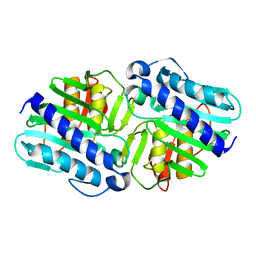 | | Crystal Structure of apo-form (P32) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | 分子名称: | fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBZ
 
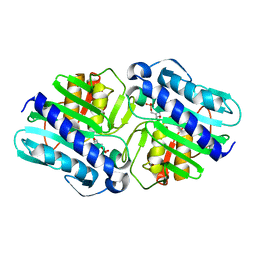 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Calcium ions and Fructose-1,6 bisphosphate | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBX
 
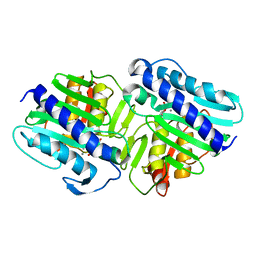 | | Crystal Structure of a ternary complex of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with Calcium ions and D-myo-Inositol-1-Phosphate | | 分子名称: | CALCIUM ION, D-MYO-INOSITOL-1-PHOSPHATE, fructose 1,6-bisphosphatase/inositol monophosphatase | | 著者 | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | 登録日 | 2002-04-04 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
5K83
 
 | | Crystal Structure of a Primate APOBEC3G N-Domain, in Complex with ssDNA | | 分子名称: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | 著者 | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | 登録日 | 2016-05-27 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K81
 
 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | 分子名称: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | 著者 | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | 登録日 | 2016-05-27 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.006 Å) | | 主引用文献 | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K82
 
 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | 分子名称: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | 著者 | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | 登録日 | 2016-05-27 | | 公開日 | 2016-08-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.913 Å) | | 主引用文献 | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5KK5
 
 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | 分子名称: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | 著者 | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | 登録日 | 2016-06-21 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.289 Å) | | 主引用文献 | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
8UD2
 
 | |
8UD3
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form | | 分子名称: | Non-structural protein 15, RNA (35-MER) | | 著者 | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.67 Å) | | 主引用文献 | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD4
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1 | | 分子名称: | Non-structural protein 15, RNA (35-MER) | | 著者 | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD5
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2 | | 分子名称: | Non-structural protein 15, RNA (35-MER) | | 著者 | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | 登録日 | 2023-09-28 | | 公開日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
5QCS
 
 | | Crystal structure of BACE complex with BMC024 | | 分子名称: | (2R,4S)-N-BUTYL-4-HYDROXY-2-METHYL- 4-((E)-(4AS,12R,15S,17AS)-15-METHYL -14,17-DIOXO-2,3,4,4A,6,9,11,12,13, 14,15,16,17,17A-TETRADECAHYDRO-1H-5 ,10-DITHIA-1,13,16-TRIAZA-BENZOCYCL OPENTADECEN-12-YL)-BUTYRAMIDE, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD4
 
 | | Crystal structure of BACE complex with BMC023 | | 分子名称: | Beta-secretase 1, {(E)-(3R,6S,9R)-3-[(1S,3R)-3-((S)-1 -BUTYLCARBAMOYL-2-METHYL-PROPYLCARB AMOYL)-1-HYDROXY-BUTYL]-6-METHYL-5, 8-DIOXO-1,11-DITHIA-4,7-DIAZA-CYCLO PENTADEC-13-EN-9-YL}-CARBAMIC ACID TERT-BUTYL ESTER | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.112 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
4PXZ
 
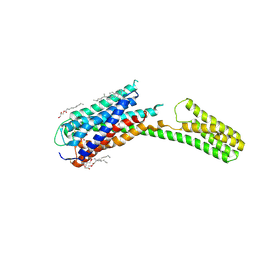 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | 著者 | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
3IO8
 
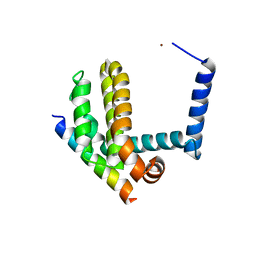 | | BimL12F in complex with Bcl-xL | | 分子名称: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | 著者 | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | 登録日 | 2009-08-14 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
3IO9
 
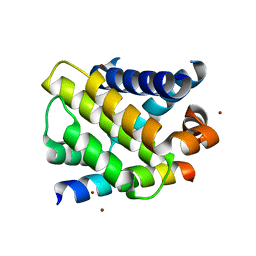 | | BimL12Y in complex with Mcl-1 | | 分子名称: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | 著者 | Czabotar, P.E, Lee, E.F, Yang, H, Sleebs, B.E, Lessene, G, Colman, P.M, Smith, B.J, Fairlie, W.D. | | 登録日 | 2009-08-14 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | 著者 | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | 登録日 | 2013-12-02 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
4PY0
 
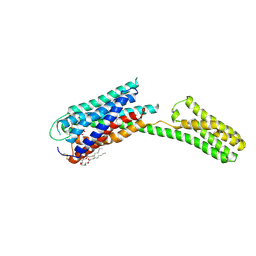 | | Crystal structure of P2Y12 receptor in complex with 2MeSATP | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), P2Y purinoceptor 12, ... | | 著者 | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
3CMT
 
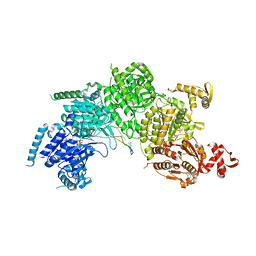 | | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*DTP*DTP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DCP*DTP*DTP*DTP*DT)-3'), DNA (5'-D(P*DGP*DGP*DTP*DGP*DGP*DG)-3'), ... | | 著者 | Chen, Z, Yang, H, Pavletich, N.P. | | 登録日 | 2008-03-24 | | 公開日 | 2008-05-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures.
Nature, 453, 2008
|
|
3GIZ
 
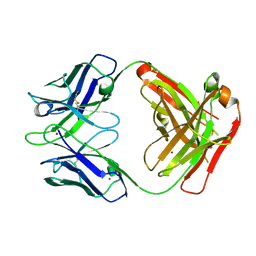 | | Crystal structure of the Fab fragment of anti-CD20 antibody Ofatumumab | | 分子名称: | Fab fragment of anti-CD20 antibody Ofatumumab, heavy chain, light chain, ... | | 著者 | Du, J, Yang, H, Ding, J. | | 登録日 | 2009-03-07 | | 公開日 | 2009-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the Fab fragment of therapeutic antibody Ofatumumab provides insights into the recognition mechanism with CD20
Mol.Immunol., 46, 2009
|
|
4FUW
 
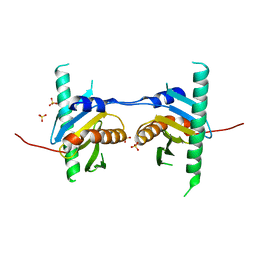 | | Crystal structure of Ego3 mutant | | 分子名称: | Protein SLM4, SULFATE ION | | 著者 | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | 登録日 | 2012-06-28 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
5QD7
 
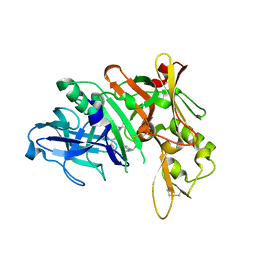 | | Crystal structure of BACE complex with BMC014 | | 分子名称: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | 著者 | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | 登録日 | 2017-12-01 | | 公開日 | 2020-06-03 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5U78
 
 | |
5U77
 
 | | Crystal structure of ORP8 PH domain | | 分子名称: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | 著者 | Ghai, R, Yang, H. | | 登録日 | 2016-12-11 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.157 Å) | | 主引用文献 | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
