6LCU
 
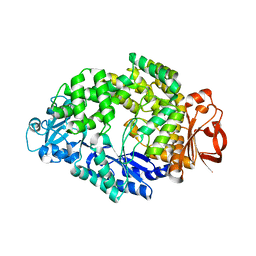 | |
6LFM
 
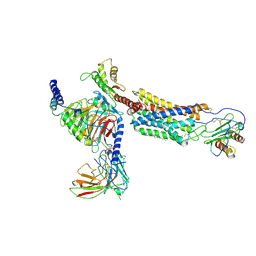 | | Cryo-EM structure of a class A GPCR | | 分子名称: | C-X-C chemokine receptor type 2, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | 登録日 | 2019-12-03 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
6LFO
 
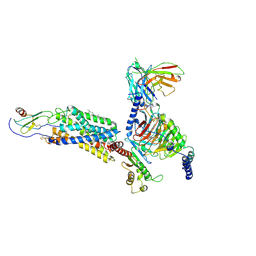 | | Cryo-EM structure of a class A GPCR monomer | | 分子名称: | C-X-C chemokine receptor type 2, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | 登録日 | 2019-12-03 | | 公開日 | 2020-09-02 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
6LFL
 
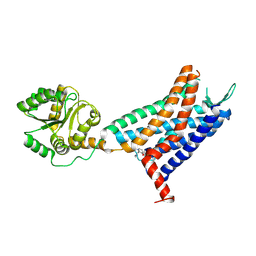 | | Crystal structure of a class A GPCR | | 分子名称: | 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, C-X-C chemokine receptor type 2,GlgA glycogen synthase,C-X-C chemokine receptor type 2 | | 著者 | Liu, Z.J, Hua, T, Liu, K.W, Wu, L.J. | | 登録日 | 2019-12-03 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of CXC chemokine receptor 2 activation and signalling.
Nature, 585, 2020
|
|
6DQ7
 
 | |
6DQ4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR GSK-J1 | | 分子名称: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-06-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.392 Å) | | 主引用文献 | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ9
 
 | | Linked KDM5A JMJ Domain Bound to the Covalent Inhibitor N69 i.e. [2-((3-acrylamidophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | 分子名称: | 1,2-ETHANEDIOL, 2-{(R)-[3-(acryloylamino)phenyl][2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2018-06-10 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.748 Å) | | 主引用文献 | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ5
 
 | |
6DQE
 
 | |
6DQD
 
 | |
7C01
 
 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | 著者 | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
6LGT
 
 | | Complex structure of HPPD with an inhibitor Y16542 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chlorophenyl)-6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | 著者 | Lin, H.Y, Yang, W.C, Yang, G.F. | | 登録日 | 2019-12-05 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.794 Å) | | 主引用文献 | Discovery of Novel Pyrazole-Quinazoline-2,4-dione Hybrids as 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 68, 2020
|
|
5B88
 
 | | RRM-like domain of DEAD-box protein, CsdA | | 分子名称: | ATP-dependent RNA helicase DeaD | | 著者 | Xu, L, Peng, J, Zhang, J, Wu, J, Tang, Y, Shi, Y. | | 登録日 | 2016-06-13 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5GJU
 
 | | DEAD-box RNA helicase | | 分子名称: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DeaD | | 著者 | Xu, L, Li, F, Wang, L, Shi, Y. | | 登録日 | 2016-07-02 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | 著者 | He, Q.W, Xu, Z.P, Xie, Y.F. | | 登録日 | 2023-01-02 | | 公開日 | 2023-12-06 | | 実験手法 | ELECTRON MICROSCOPY (2.36 Å) | | 主引用文献 | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
8HWT
 
 | |
4GVJ
 
 | | Tyk2 (JH1) in complex with adenosine di-phosphate | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Non-receptor tyrosine-protein kinase TYK2 | | 著者 | Liang, J, Abbema, A.V, Bao, L, Barrett, K, Beresini, M, Berezhkovskiy, L, Blair, W, Chang, C, Driscoll, J, Eigenbrot, C, Ghilardi, N, Gibbons, P, Halladay, J, Johnson, A, Kohli, P.B, Lai, Y, Liimatta, M, Mantik, P, Menghrajani, K, Murray, J, Sambrone, A, Shao, Y, Shia, S, Shin, Y, Smith, J, Sohn, S, Stanley, M, Tsui, V, Ultsch, M, Wu, L, Zhang, B, Magnuson, S. | | 登録日 | 2012-08-30 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Lead identification of novel and selective TYK2 inhibitors.
Eur.J.Med.Chem., 67, 2013
|
|
5H65
 
 | | Crystal structure of human POT1 and TPP1 | | 分子名称: | Adrenocortical dysplasia protein homolog, Protection of telomeres protein 1, ZINC ION | | 著者 | Chen, C, Wu, J, Lei, M. | | 登録日 | 2016-11-10 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer.
Nat Commun, 8, 2017
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-08-18 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | 登録日 | 2022-08-19 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
5ZTY
 
 | | Crystal structure of human G protein coupled receptor | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Li, X.T, Hua, T, Wu, L.J, Liu, Z.J. | | 登録日 | 2018-05-05 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Human Cannabinoid Receptor CB2
Cell, 176, 2019
|
|
6S00
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid | | 分子名称: | GLYCEROL, N-acetyl-beta-neuraminic acid, TETRAETHYLENE GLYCOL, ... | | 著者 | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | 登録日 | 2019-06-13 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6S0E
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ACETATE ION, ... | | 著者 | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | 登録日 | 2019-06-14 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6S0F
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | 著者 | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | 登録日 | 2019-06-14 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
