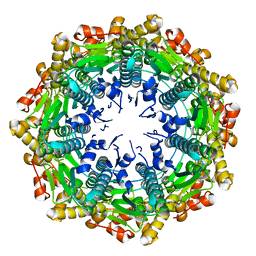
4EMM
 
 | |
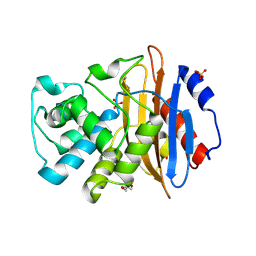
5UL8
 
 | |
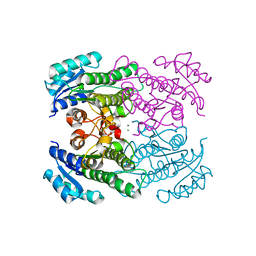
4RF4
 
 | |
5UJ4
 
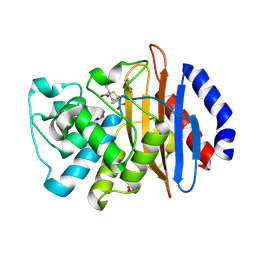 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed faropenem | | 分子名称: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | 著者 | Pemberton, O.A, Chen, Y. | | 登録日 | 2017-01-16 | | 公開日 | 2017-04-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
8K31
 
 | | The complex of WRKY33 C terminal DBD and SIB1 | | 分子名称: | Probable WRKY transcription factor 33, Sigma factor binding protein 1, chloroplastic, ... | | 著者 | Dong, X, Gong, Z, Hu, Y.F. | | 登録日 | 2023-07-14 | | 公開日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1.
Commun Biol, 7, 2024
|
|
7BST
 
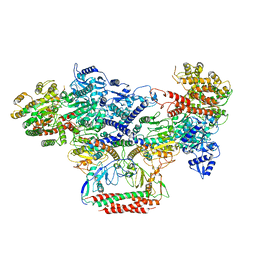 | | EcoR124I-Ocr in the Intermediate State | | 分子名称: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | 著者 | Gao, Y, Gao, P. | | 登録日 | 2020-03-31 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.37 Å) | | 主引用文献 | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BTR
 
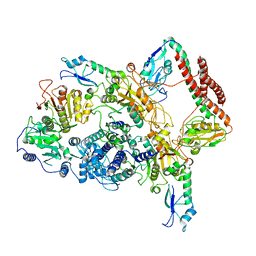 | | EcoR124I-ArdA in the Restriction-Alleviation State | | 分子名称: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | 著者 | Gao, Y, Gao, P. | | 登録日 | 2020-04-02 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.54 Å) | | 主引用文献 | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
7BX7
 
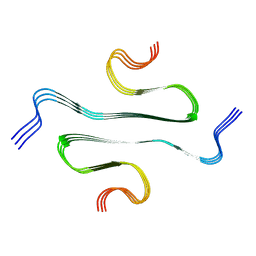 | |
5EDS
 
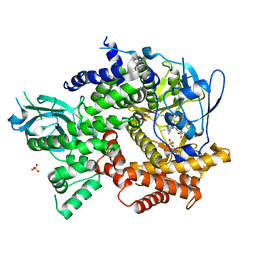 | | Crystal structure of human PI3K-gamma in complex with benzimidazole inhibitor 5 | | 分子名称: | 4-azanyl-6-[[(1~{S})-1-[6-fluoranyl-1-(3-methylsulfonylphenyl)benzimidazol-2-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2015-10-21 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery, Optimization, and in Vivo Evaluation of Benzimidazole Derivatives AM-8508 and AM-9635 as Potent and Selective PI3K delta Inhibitors.
J.Med.Chem., 59, 2016
|
|
8TAB
 
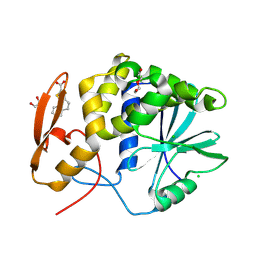 | | RTA-PD00589 | | 分子名称: | 1,2-ETHANEDIOL, 4H,5H-naphtho[1,2-b]thiophene-2-carboxylic acid, CHLORIDE ION, ... | | 著者 | Rudolph, M.J, Tumer, N. | | 登録日 | 2023-06-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structure-based design and optimization of a new class of small molecule inhibitors targeting the P-stalk binding pocket of ricin.
Bioorg.Med.Chem., 100, 2024
|
|
8T9V
 
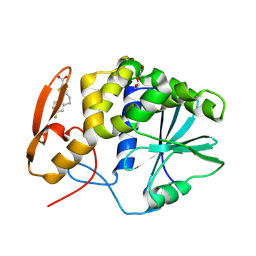 | | RTA-RUNT-59 complex structure | | 分子名称: | (9aP)-7-fluoro-4,5-dihydronaphtho[1,2-b]thiophene-2-carboxylic acid, NONAETHYLENE GLYCOL, Ricin | | 著者 | Rudolph, M.J, Tumer, N. | | 登録日 | 2023-06-26 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.945 Å) | | 主引用文献 | Structure-based design and optimization of a new class of small molecule inhibitors targeting the P-stalk binding pocket of ricin.
Bioorg.Med.Chem., 100, 2024
|
|
8TAD
 
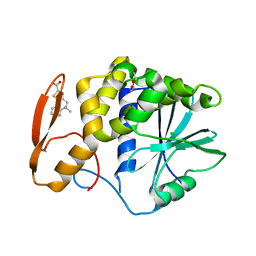 | | RTA in complex with inhibitor RUNT-206 | | 分子名称: | (9aM)-5,5-dimethyl-4,5-dihydronaphtho[1,2-b]thiophene-2-carboxylic acid, CHLORIDE ION, NONAETHYLENE GLYCOL, ... | | 著者 | Rudolph, M.J, Tumer, N. | | 登録日 | 2023-06-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structure-based design and optimization of a new class of small molecule inhibitors targeting the P-stalk binding pocket of ricin.
Bioorg.Med.Chem., 100, 2024
|
|
7A3O
 
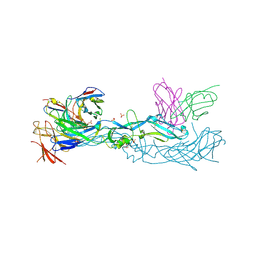 | | Crystal structure of dengue 1 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | 分子名称: | Core protein, GLYCEROL, SULFATE ION, ... | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3R
 
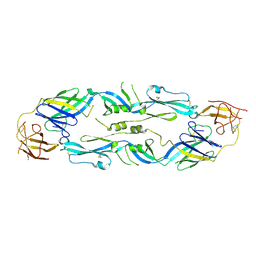 | | Crystal structure of dengue 1 virus envelope glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3N
 
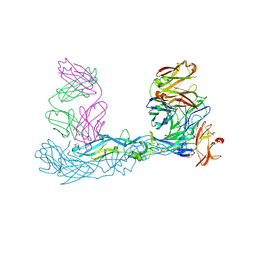 | | Crystal structure of Zika virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C10 | | 分子名称: | CALCIUM ION, Core protein, EDE1 C10 Fab | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3T
 
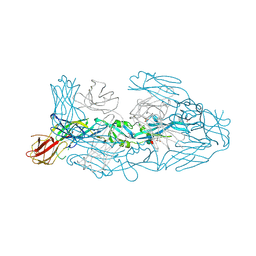 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | 分子名称: | Core protein, EDE1 C8 antibody Fab fragment, GLYCEROL, ... | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3U
 
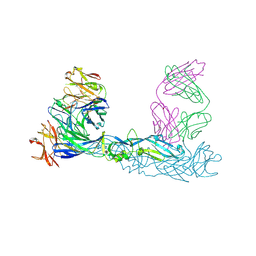 | | Crystal structure of Zika virus envelope glycoprotein in complex with the divalent F(ab')2 fragment of the broadly neutralizing human antibody EDE1 C10 | | 分子名称: | EDE1 C10 antibody divalent F(ab')2 fragment, EDE1 C10 divalent F(ab')2 fragment, Envelope protein | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3P
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Single Chain Variable Fragment | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3S
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, SULFATE ION | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Navarro-Sanchez, E, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3Q
 
 | | Crystal structure of dengue 4 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | 分子名称: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | 著者 | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | 登録日 | 2020-08-18 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
4RPA
 
 | |
5U6J
 
 | | Factor VIIa in complex with the inhibitor 3-{[(2R)-17-ethyl-4-methyl-3,12-dioxo-7-[(propan-2-yl)sulfonyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-2-yl]amino}benzamide | | 分子名称: | 3-{[(2R)-17-ethyl-4-methyl-3,12-dioxo-7-[(propan-2-yl)sulfonyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-2-yl]amino}benzamide, CALCIUM ION, Coagulation factor VII Heavy Chain, ... | | 著者 | Wei, A. | | 登録日 | 2016-12-08 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Neutral macrocyclic factor VIIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5HQC
 
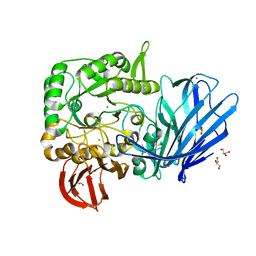 | | A Glycoside Hydrolase Family 97 enzyme R171K variant from Pseudoalteromonas sp. strain K8 | | 分子名称: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Li, J, He, C, Xiao, Y. | | 登録日 | 2016-01-21 | | 公開日 | 2017-01-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Structures of PspAG97A alpha-glucoside hydrolase reveal a novel mechanism for chloride induced activation.
J. Struct. Biol., 196, 2016
|
|
1YF3
 
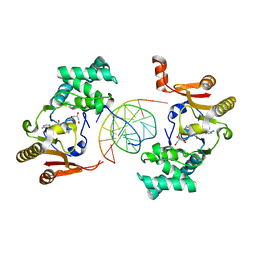 | | T4Dam in Complex with AdoHcy and 13-mer Oligonucleotide Making Non- and Semi-specific (~1/4) Contact | | 分子名称: | 5'-D(*AP*CP*CP*AP*TP*GP*AP*TP*CP*TP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*CP*AP*TP*GP*G)-3', DNA adenine methylase, ... | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2004-12-30 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YFL
 
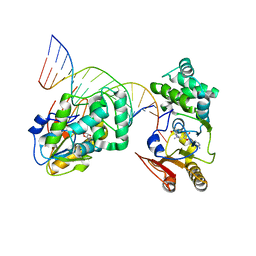 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | 分子名称: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | 著者 | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | 登録日 | 2005-01-03 | | 公開日 | 2005-05-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
