2OSL
 
 | | Crystal structure of Rituximab Fab in complex with an epitope peptide | | 分子名称: | B-lymphocyte antigen CD20, heavy chain of the Rituximab Fab fragment,heavy chain of the Rituximab Fab fragment, light chain of the Rituximab Fab fragment,light chain of the Rituximab Fab fragment | | 著者 | Du, J, Zhong, C, Ding, J. | | 登録日 | 2007-02-06 | | 公開日 | 2007-04-10 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for recognition of CD20 by therapeutic antibody Rituximab
J.Biol.Chem., 282, 2007
|
|
7DA4
 
 | | Cryo-EM structure of amyloid fibril formed by human RIPK3 | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Zhao, K, Ma, Y.Y, Sun, Y.P, Li, D, Liu, C. | | 登録日 | 2020-10-14 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3JZS
 
 | |
3JZO
 
 | |
3JZQ
 
 | |
3JZR
 
 | |
3JZP
 
 | |
5XTJ
 
 | | Mannanase(RmMan134A) | | 分子名称: | Endo beta-1,4-mannanase | | 著者 | Jiang, Z.Q, You, X, Huang, P. | | 登録日 | 2017-06-19 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XUG
 
 | | Complex structure(RmMan134A-M5). | | 分子名称: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-06-23 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XU5
 
 | | Complex structure of RmMan134A-M4 | | 分子名称: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-06-22 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XTT
 
 | | Crystal structure of RmMan134A-M3 complex | | 分子名称: | beta-1,4-mannanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-06-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5XUL
 
 | | Complex structure (RmMan134A-M6). | | 分子名称: | beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, endo-1,4-beta-mannanase | | 著者 | Jiang, Z.Q, You, X, Yang, S.Q, Huang, P, Ma, J.W. | | 登録日 | 2017-06-23 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a novel glycoside hydrolase family 134 beta-mannanase.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
3KA0
 
 | |
3KC3
 
 | | MK2 complexed to inhibitor N4-(7-(benzofuran-2-yl)-1H-indazol-5-yl)pyrimidine-2,4-diamine | | 分子名称: | MAP kinase-activated protein kinase 2, N~4~-[7-(1-benzofuran-2-yl)-1H-indazol-5-yl]pyrimidine-2,4-diamine | | 著者 | Argiriadi, M.A, Talanian, R.V, Borhani, D.W. | | 登録日 | 2009-10-20 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | 2,4-Diaminopyrimidine MK2 inhibitors. Part I: Observation of an unexpected inhibitor binding mode.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5WC2
 
 | | Crystal Structure of ADP-bound human TRIP13 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | 著者 | Jeong, B.-C, Luo, X. | | 登録日 | 2017-06-29 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
7R9X
 
 | |
3V19
 
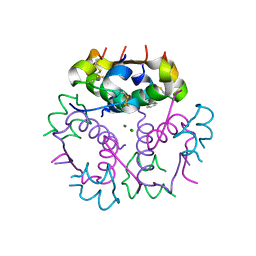 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | 分子名称: | CHLORIDE ION, Insulin, PHENOL, ... | | 著者 | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | 登録日 | 2011-12-09 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
7YI4
 
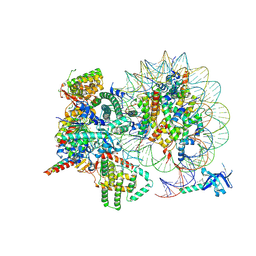 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | 分子名称: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
3V1G
 
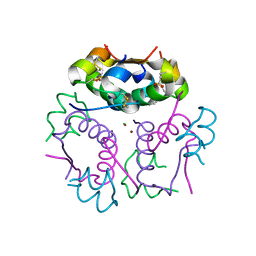 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | 分子名称: | CHLORIDE ION, Insulin, PHENOL, ... | | 著者 | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | 登録日 | 2011-12-09 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
7YI2
 
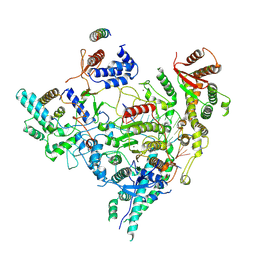 | | Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex | | 分子名称: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI3
 
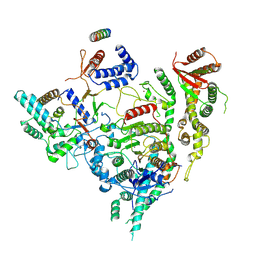 | | Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex | | 分子名称: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI1
 
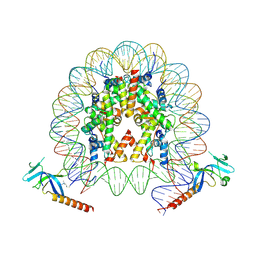 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | 分子名称: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI0
 
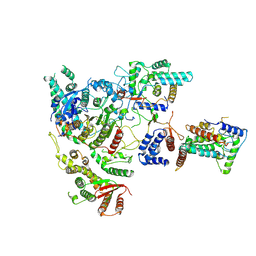 | | Cryo-EM structure of Rpd3S complex | | 分子名称: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI5
 
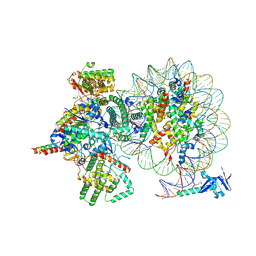 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | 分子名称: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
4YGY
 
 | |
