3B95
 
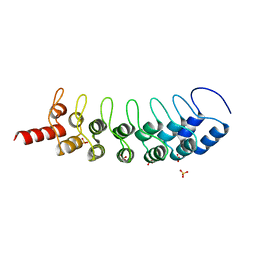 | |
7WA9
 
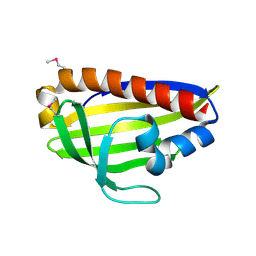 | |
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | 分子名称: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | 著者 | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2016-09-29 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.755 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5GWE
 
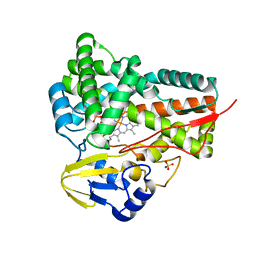 | | cytochrome P450 CREJ | | 分子名称: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Dong, S, liu, X, Wang, X, Feng, Y. | | 登録日 | 2016-09-11 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7XCK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with S309 fab (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Xie, Y.F, Liu, S. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCI
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron RBD in complex with human ACE2 ectodomain (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCH
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (two-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
7XCO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with S309 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 Fab heavy chain, ... | | 著者 | Gao, G.F, Qi, J.X, Zhao, Z.N, Liu, S, Xie, Y.F. | | 登録日 | 2022-03-24 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | 分子名称: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | 著者 | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | 登録日 | 2014-04-16 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | 分子名称: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | 著者 | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | 登録日 | 2016-09-29 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
7RTB
 
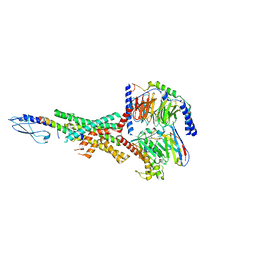 | | Peptide-19 bound to the Glucagon-Like Peptide-1 Receptor (GLP-1R) | | 分子名称: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Johnson, R.M, Danev, R, Sexton, P.M, Wootten, D. | | 登録日 | 2021-08-12 | | 公開日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (2.14 Å) | | 主引用文献 | Cryo-EM structure of the dual incretin receptor agonist, peptide-19, in complex with the glucagon-like peptide-1 receptor.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
7XLB
 
 | |
7YA1
 
 | | Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7YAD
 
 | | Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, S309 neutralizing antibody light chain, ... | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, F. | | 登録日 | 2022-06-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9S
 
 | | Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | 登録日 | 2022-06-26 | | 公開日 | 2022-08-31 | | 最終更新日 | 2022-09-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7Y9Z
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Gao, G.F, Qi, J.X, Liu, S, Zhao, Z.N. | | 登録日 | 2022-06-26 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape.
Nat Commun, 13, 2022
|
|
7E3J
 
 | |
6BH3
 
 | |
6BGX
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | 分子名称: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.882 Å) | | 主引用文献 | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGY
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | 分子名称: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
8D4J
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4N
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
6BH1
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (S)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N52) | | 分子名称: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.932 Å) | | 主引用文献 | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | 分子名称: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | 著者 | Sacco, M, Ma, C, Wang, J, Chen, Y. | | 登録日 | 2020-08-12 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
6BGV
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | 分子名称: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.592 Å) | | 主引用文献 | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
