6WQZ
 
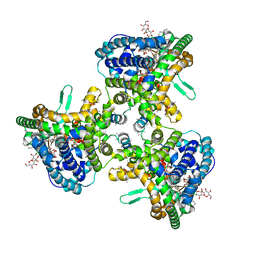 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | 分子名称: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | 著者 | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | 登録日 | 2020-04-29 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
5YKI
 
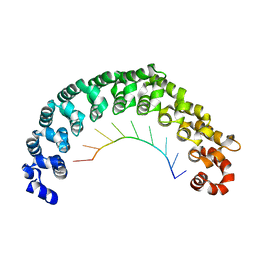 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | 分子名称: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | 著者 | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | 登録日 | 2017-10-14 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
5C8C
 
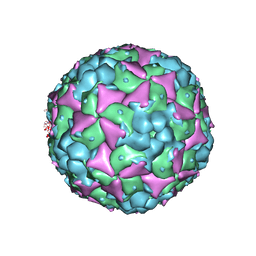 | | Crystal structure of recombinant coxsackievirus A16 capsid | | 分子名称: | CHLORIDE ION, POTASSIUM ION, STEARIC ACID, ... | | 著者 | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | 登録日 | 2015-06-25 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5YKH
 
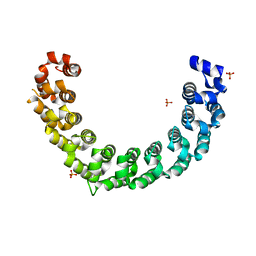 | | Crystal structure of the engineered nine-repeat PUF domain | | 分子名称: | PHOSPHATE ION, Pumilio homolog 1 | | 著者 | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | 登録日 | 2017-10-14 | | 公開日 | 2018-03-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.457 Å) | | 主引用文献 | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | 分子名称: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.57150865 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | 分子名称: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.20017123 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | 分子名称: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | 著者 | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65004492 Å) | | 主引用文献 | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
4WF1
 
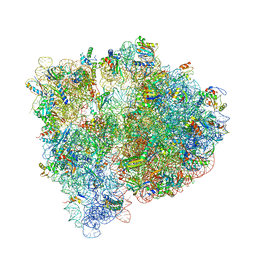 | | Crystal structure of the E. coli ribosome bound to negamycin. | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | 登録日 | 2014-09-11 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6WR4
 
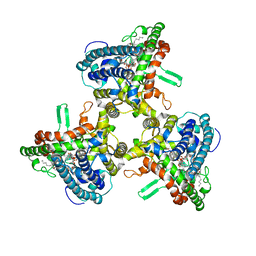 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | 分子名称: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | 著者 | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | 登録日 | 2020-04-29 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
6WMQ
 
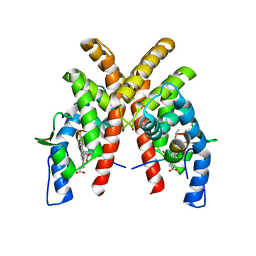 | |
6WMS
 
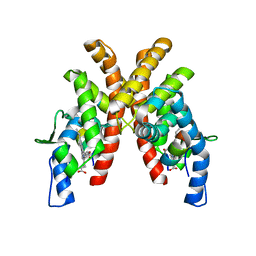 | |
4GAZ
 
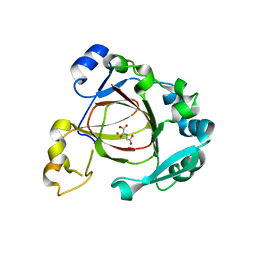 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | 分子名称: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | 著者 | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | 登録日 | 2012-07-26 | | 公開日 | 2013-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
6GYU
 
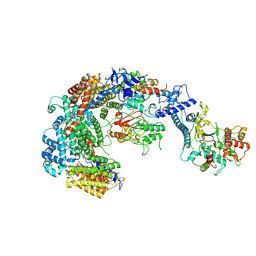 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5CTW
 
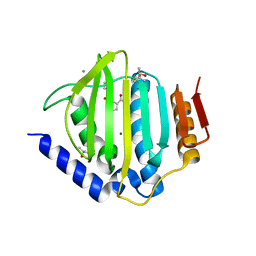 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTX
 
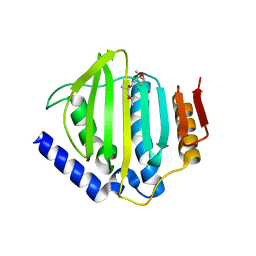 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-21 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8WZP
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | 分子名称: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | 著者 | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | 登録日 | 2023-11-02 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
5CTY
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | 著者 | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | 登録日 | 2015-07-24 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8FTL
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1 | | 分子名称: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-2-chloro-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]acetamide | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2023-01-12 | | 公開日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of the SARS-CoV-2 (COVID-19) main protease (Mpro) in complex with inhibitor Jun89-3-C1
To Be Published
|
|
5ZTE
 
 | | Crystal structure of PrxA C119S mutant from Arabidopsis thaliana | | 分子名称: | 2-Cys peroxiredoxin BAS1, chloroplastic | | 著者 | Yang, Y, Cai, W, Wang, J, Pan, W, Liu, L, Wang, M, Zhang, M. | | 登録日 | 2018-05-03 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of Arabidopsis thaliana peroxiredoxin A C119S mutant.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2MOX
 
 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | 分子名称: | Sorbin and SH3 domain-containing protein 1 | | 著者 | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | 登録日 | 2014-05-07 | | 公開日 | 2014-05-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
6GYP
 
 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | 分子名称: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYS
 
 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4XHS
 
 | | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction | | 分子名称: | FORMIC ACID, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 12, ... | | 著者 | Jin, T, Huang, M, Jiang, J, Xiao, T. | | 登録日 | 2015-01-06 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction
To Be Published
|
|
