6WOT
 
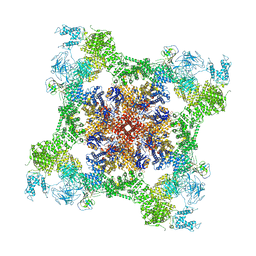 | | Cryo-EM structure of recombinant rabbit Ryanodine Receptor type 1 mutant R164C in complex with FKBP12.6 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | 著者 | Iyer, K.A, Hu, Y, Kurebayashi, N, Murayama, T, Samso, M. | | 登録日 | 2020-04-25 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM.
Sci Adv, 6, 2020
|
|
7JMB
 
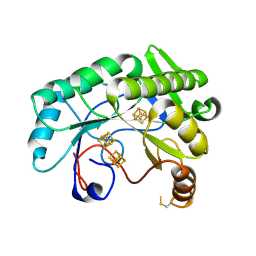 | | Crystal structure of Nitrogenase iron-molybdenum cofactor biosynthesis enzyme NifB from Methanothermobacter thermautotrophicus with three Fe4S4 clusters | | 分子名称: | IRON/SULFUR CLUSTER, Nitrogenase iron-molybdenum cofactor biosynthesis protein NifB | | 著者 | Kang, W, Rettberg, L, Ribbe, M.W, Hu, Y. | | 登録日 | 2020-07-31 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-Ray Crystallographic Analysis of NifB with a Full Complement of Clusters: Structural Insights into the Radical SAM-Dependent Carbide Insertion During Nitrogenase Cofactor Assembly.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7JMA
 
 | |
2P0P
 
 | |
8UUI
 
 | | X-ray structure of Interleukin-23 in complex with peptide 23-446 | | 分子名称: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Joseph, J.S, Sridhar, V.S, Liu, M, Hu, Y, Gatchalian, J. | | 登録日 | 2023-11-01 | | 公開日 | 2025-07-09 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Identification of an Induced Orthosteric Pocket in IL-23: A New Avenue for Non-biological Therapeutic Targeting.
Acs Chem.Biol., 2025
|
|
3VNI
 
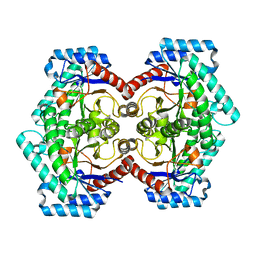 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | 分子名称: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNJ
 
 | | Crystal structures of D-Psicose 3-epimerase with D-psicose from Clostridium cellulolyticum H10 | | 分子名称: | D-psicose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
4KT8
 
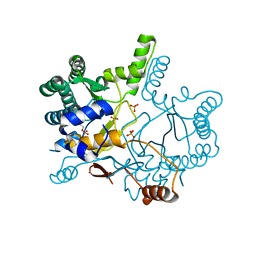 | | The complex structure of Rv3378c-Y51FY90F with substrate, TPP | | 分子名称: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | 著者 | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | 登録日 | 2013-05-20 | | 公開日 | 2014-02-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
3VNM
 
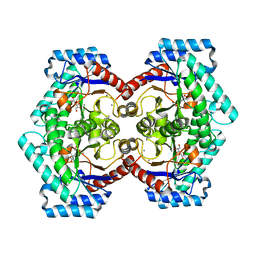 | | Crystal structures of D-Psicose 3-epimerase with D-sorbose from Clostridium cellulolyticum H10 | | 分子名称: | D-sorbose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-17 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | 分子名称: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNK
 
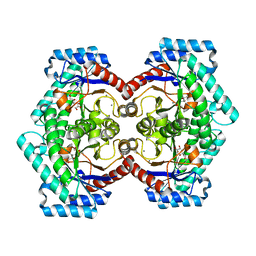 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | 分子名称: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
2MN6
 
 | |
7T64
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | 著者 | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T65
 
 | | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the open state | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | 著者 | Iyer, K.A, Hu, Y, Murayama, T, Samso, M. | | 登録日 | 2021-12-13 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2MI2
 
 | |
2LZ5
 
 | | Solution structure of a Novel Alpha-Conotoxin TxIB | | 分子名称: | Conotoxin_TxIB | | 著者 | Luo, S, Zhangsun, D, Wu, Y, Zhu, X, Hu, Y, McIntyre, M, Christensen, S, Akcan, M, Craik, D, McIntosh, J. | | 登録日 | 2012-09-23 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-11-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Characterization of a novel alpha-conotoxin from conus textile that selectively targets alpha6/alpha3beta2beta3 nicotinic acetylcholine receptors.
J.Biol.Chem., 288, 2013
|
|
2M3I
 
 | | Characterization of a Novel Alpha4/6-Conotoxin TxIC from Conus textile that Potently Blocks alpha3beta4 Nicotinic Acetylcholine Receptors | | 分子名称: | Alpha-conotoxin | | 著者 | Luo, S, Zhangsun, D, Zhu, X, Wu, Y, Hu, Y, Christensen, S, Akcan, M, Craik, D.J, McIntosh, J.M. | | 登録日 | 2013-01-20 | | 公開日 | 2013-12-04 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Characterization of a Novel alpha-Conotoxin TxID from Conus textile That Potently Blocks Rat alpha 3 beta 4 Nicotinic Acetylcholine Receptors.
J.Med.Chem., 56, 2013
|
|
2MN7
 
 | |
6UG0
 
 | | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Hu, Y, Ribbe, M.W. | | 登録日 | 2019-09-25 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
4XIY
 
 | | Crystal structure of ketol-acid reductoisomerase from Azotobacter | | 分子名称: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Ketol-acid reductoisomerase, ... | | 著者 | Spatzal, T, Cahn, J.K.B, Wiig, J.A, Einsle, O, Hu, Y, Ribbe, M.W, Arnold, F.H. | | 登録日 | 2015-01-07 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
3W02
 
 | | Crystal structure of PcrB complexed with SO4 from Staphylococcus aureus subsp. aureus Mu3 | | 分子名称: | Heptaprenylglyceryl phosphate synthase, SULFATE ION | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3W01
 
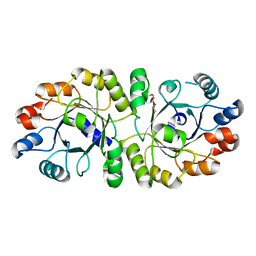 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | 分子名称: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Hu, Y, Ribbe, M.W. | | 登録日 | 2020-02-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
3VZZ
 
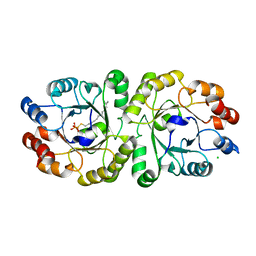 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | 分子名称: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VZX
 
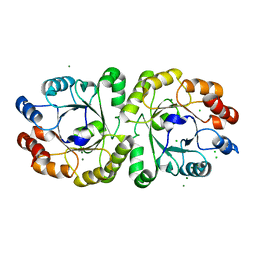 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | 分子名称: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | 著者 | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
