5WWN
 
 | | Crystal structure of Tsr1 | | 分子名称: | Ribosome biogenesis protein TSR1, SULFATE ION | | 著者 | Ye, K, Wang, B. | | 登録日 | 2017-01-03 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5WXM
 
 | | Crystal structure of the Imp3 and Mpp10 complex | | 分子名称: | SULFATE ION, U3 small nucleolar RNA-associated protein MPP10, U3 small nucleolar ribonucleoprotein protein IMP3 | | 著者 | Ye, K, Zheng, S. | | 登録日 | 2017-01-07 | | 公開日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
5IH9
 
 | | MELK in complex with NVS-MELK8A | | 分子名称: | 1-methyl-4-[4-(4-{3-[(piperidin-4-yl)methoxy]pyridin-4-yl}-1H-pyrazol-1-yl)phenyl]piperazine, Maternal embryonic leucine zipper kinase | | 著者 | Sprague, E.R, Puleo, D.E. | | 登録日 | 2016-02-29 | | 公開日 | 2016-06-01 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
5WWO
 
 | | Crystal structure of Enp1 | | 分子名称: | Essential nuclear protein 1, Protein LTV1 | | 著者 | Ye, K, Zhang, W. | | 登録日 | 2017-01-03 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
2Z4D
 
 | |
4OAS
 
 | | co-crystal structure of MDM2 (17-111) in complex with compound 25 | | 分子名称: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-(tert-butylsulfonyl)butan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetic acid | | 著者 | Huang, X. | | 登録日 | 2014-01-06 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of AMG 232, a Potent, Selective, and Orally Bioavailable MDM2-p53 Inhibitor in Clinical Development.
J.Med.Chem., 57, 2014
|
|
8DEA
 
 | |
8DG6
 
 | |
8D95
 
 | |
8W9B
 
 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-8557 binding at an allosteric site | | 分子名称: | 1-[(1S)-1-(5-fluoranyl-3-methyl-1-benzofuran-2-yl)-2-methyl-propyl]-3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)urea, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Huang, X, Ren, X, Zhong, W. | | 登録日 | 2023-09-05 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
8W9A
 
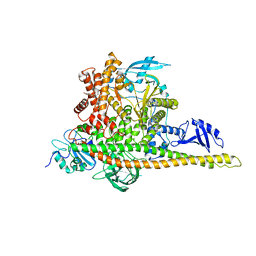 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | 分子名称: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Huang, X, Ren, X, Zhong, W. | | 登録日 | 2023-09-05 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
4ZOL
 
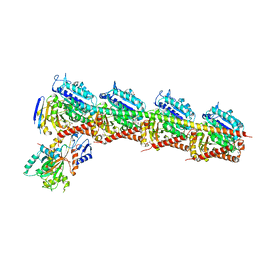 | | Crystal Structure of Tubulin-Stathmin-TTL-Tubulysin M Complex | | 分子名称: | (2R,4R)-4-{[(2-{(1R,3R)-1-(acetyloxy)-4-methyl-3-[methyl(N-{[(2S)-1-methylpiperidin-2-yl]carbonyl}-D-isoleucyl)amino]pentyl}-1,3-thiazol-4-yl)carbonyl]amino}-2-methyl-5-phenylpentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Wang, Y, Zhang, R. | | 登録日 | 2015-05-06 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.919 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
4YVQ
 
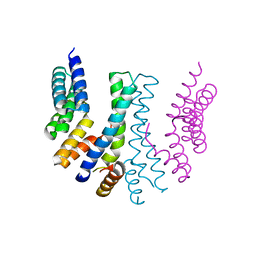 | |
4ZHQ
 
 | |
4ZI7
 
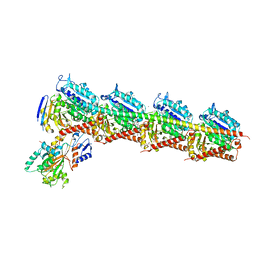 | |
3CWG
 
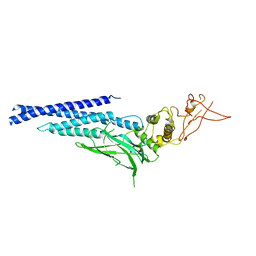 | | Unphosphorylated mouse STAT3 core fragment | | 分子名称: | Signal transducer and activator of transcription 3 | | 著者 | Ren, Z, Mao, X, Mertens, C, Krishnaraj, R, Qin, J, Mandal, P.K, Romanowshi, M.J, McMurray, J.S. | | 登録日 | 2008-04-21 | | 公開日 | 2008-07-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of unphosphorylated STAT3 core fragment.
Biochem.Biophys.Res.Commun., 374, 2008
|
|
5YKO
 
 | |
1KBR
 
 | |
1TMJ
 
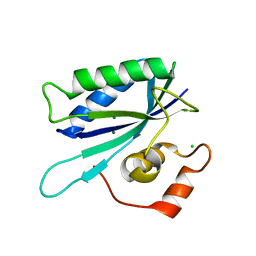 | | Crystal structure of E.coli apo-HPPK(W89A) at 1.45 Angstrom resolution | | 分子名称: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION, MAGNESIUM ION | | 著者 | Blaszczyk, J, Ji, X. | | 登録日 | 2004-06-10 | | 公開日 | 2005-06-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Is the Critical Role of Loop 3 of Escherichia coli 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase in Catalysis Due to Loop-3 Residues Arginine-84 and Tryptophan-89? Site-Directed Mutagenesis, Biochemical, and Crystallographic Studies.
Biochemistry, 44, 2005
|
|
6MQE
 
 | |
6MQS
 
 | |
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | 著者 | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | 登録日 | 2022-08-25 | | 公開日 | 2023-06-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
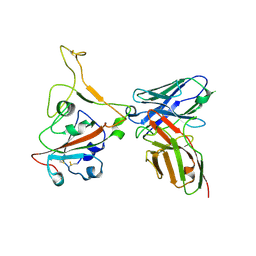 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | 分子名称: | Spike protein S1, scFv | | 著者 | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | 登録日 | 2022-08-27 | | 公開日 | 2023-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
