7E8J
 
 | |
3K8S
 
 | | Crystal Structure of PPARg in complex with T2384 | | 分子名称: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | 著者 | Wang, Z. | | 登録日 | 2009-10-14 | | 公開日 | 2009-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
5ICT
 
 | |
5YOF
 
 | |
5YOD
 
 | |
7VYQ
 
 | | Short chain dehydrogenase (SCR) cryoEM structure with NADP and ethyl 4-chloroacetoacetate | | 分子名称: | Carbonyl Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl 4-chloranyl-3-oxidanylidene-butanoate | | 著者 | Li, Y.H, Zhang, R.Z, Wang, C, Forouhar, F, Clarke, O, Vorobiev, S, Singh, S, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2021-11-15 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7EXC
 
 | | Crystal structure of T2R-TTL-1129A2 complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Yang, J.H, Yan, W. | | 登録日 | 2021-05-26 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure-Based Design and Synthesis of N-Substituted 3-Amino-beta-Carboline Derivatives as Potent alpha beta-Tubulin Degradation Agents
J.Med.Chem., 65, 2022
|
|
7DMG
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADP | | 分子名称: | (S)-specific carbonyl reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-12-03 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
6LYP
 
 | | Cryo-EM structure of AtMSL1 wild type | | 分子名称: | Mechanosensitive ion channel protein 1, mitochondrial | | 著者 | Sun, L. | | 登録日 | 2020-02-15 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural Insights into a Plant Mechanosensitive Ion Channel MSL1.
Cell Rep, 30, 2020
|
|
7DLM
 
 | | Short chain dehydrogenase (SCR) crystal structure with NADPH | | 分子名称: | Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLD
 
 | | Crystal structures of (S)-carbonyl reductases from Candida parapsilosis in different oligomerization states | | 分子名称: | Carbonyl Reductase, MAGNESIUM ION | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-27 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DN1
 
 | | Hetero-oligomers of SCR-SCR2 crystal structure with NADPH | | 分子名称: | (S)-specific carbonyl reductase, Carbonyl Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-12-08 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
7DLL
 
 | | Short chain dehydrogenase 2 (SCR2) crystal structure with NADPH | | 分子名称: | (S)-specific carbonyl reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Li, Y.H, Zhang, R.Z, Forouhar, F, Wang, C, Montelione, G.T, Szyperski, T, Xu, Y, Hunt, J.F. | | 登録日 | 2020-11-28 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Oligomeric interactions maintain active-site structure in a noncooperative enzyme family.
Embo J., 41, 2022
|
|
5DTB
 
 | |
5EHM
 
 | |
5EHS
 
 | |
5H6V
 
 | |
4FAA
 
 | |
4FA7
 
 | |
8JYW
 
 | |
8JYV
 
 | |
8ZSB
 
 | |
8ZSZ
 
 | |
8FR8
 
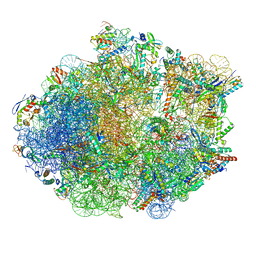 | | Structure of Mycobacterium smegmatis Rsh bound to a 70S translation initiation complex | | 分子名称: | 16S rRNA (1511-MER), 23S rRNA (3119-MER), 30S ribosomal protein S10, ... | | 著者 | Majumdar, S, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | 登録日 | 2023-01-06 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Starvation sensing by mycobacterial RelA/SpoT homologue through constitutive surveillance of translation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7C6C
 
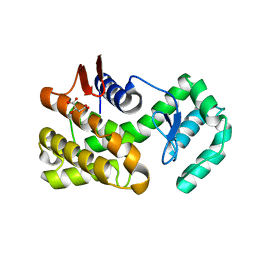 | | Crystal structure of native chitosanase from Bacillus subtilis MY002 | | 分子名称: | (2S)-2-hydroxybutanedioic acid, Chitosanase | | 著者 | Gou, Y, Liu, Z.C, Xie, T, Wang, G.G. | | 登録日 | 2020-05-21 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.258 Å) | | 主引用文献 | Structure-based rational design of chitosanase CsnMY002 for high yields of chitobiose.
Colloids Surf B Biointerfaces, 202, 2021
|
|
