6M0J
 
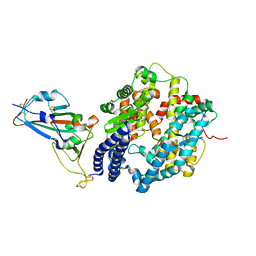 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | 著者 | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | 登録日 | 2020-02-21 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
6M2B
 
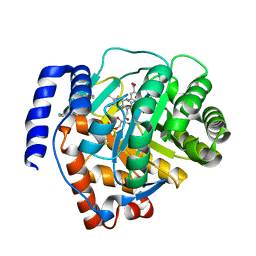 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | 分子名称: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | 著者 | Zhu, L, Li, H. | | 登録日 | 2020-02-27 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
4XXI
 
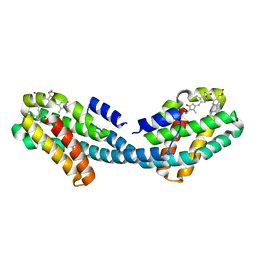 | | Crystal structure of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | 分子名称: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | 著者 | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | 登録日 | 2015-01-30 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XXU
 
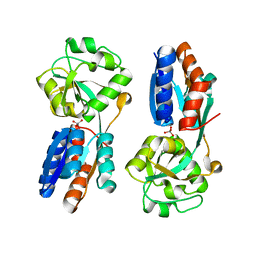 | | ModA - chromate bound | | 分子名称: | Chromate, Molybdate-binding periplasmic protein | | 著者 | He, C, Karpus, J, Ajiboye, I. | | 登録日 | 2015-01-30 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Structure of chromate bound ModA from E.coli at 1.43 Angstroms resolution.
To Be Published
|
|
4XXK
 
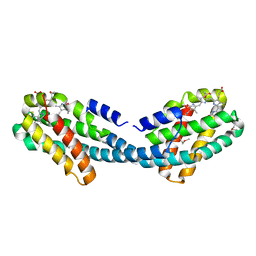 | | Crystal structure of the Semet-derivative of the Bilin-binding domain of phycobilisome core-membrane linker ApcE | | 分子名称: | PHYCOCYANOBILIN, Phycobiliprotein ApcE | | 著者 | Tang, K, Ding, W.-L, Hoppner, A, Gartner, W, Zhao, K.-H. | | 登録日 | 2015-01-30 | | 公開日 | 2015-12-16 | | 最終更新日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YLA
 
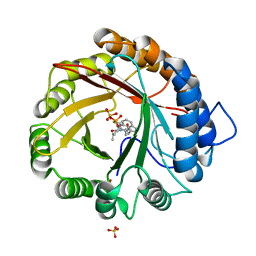 | | Crystal structure of the indole prenyltransferase MpnD complexed with indolactam V and DMSPP | | 分子名称: | (2S,5S)-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, Aromatic prenyltransferase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | 著者 | Mori, T, Morita, H, Abe, I. | | 登録日 | 2015-03-05 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases.
Nat Commun, 7, 2016
|
|
4YL7
 
 | |
4YZK
 
 | |
3CE3
 
 | |
4HND
 
 | | Crystal structure of the catalytic domain of Selenomethionine substituted human PI4KIIalpha in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | 著者 | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | 登録日 | 2012-10-19 | | 公開日 | 2014-04-09 | | 最終更新日 | 2016-12-28 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
4HNE
 
 | | Crystal structure of the catalytic domain of human type II alpha Phosphatidylinositol 4-kinase (PI4KIIalpha) in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha | | 著者 | Zhou, Q, Zhai, Y, Zhang, K, Chen, C, Sun, F. | | 登録日 | 2012-10-19 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Molecular insights into the membrane-associated phosphatidylinositol 4-kinase II alpha.
Nat Commun, 5, 2014
|
|
7SSM
 
 | |
6T5B
 
 | | KRasG12C ligand complex | | 分子名称: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Phillips, C. | | 登録日 | 2019-10-15 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6T5U
 
 | | KRasG12C ligand complex | | 分子名称: | 1-[(7R)-16-chloro-15-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,12-triazatetracyclo[8.8.0.02,7.013,18]octadeca-1(10),11,13,15,17-pentaen-5-yl]prop-2-en-1-one, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Phillips, C. | | 登録日 | 2019-10-17 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6T5V
 
 | | KRasG12C ligand complex | | 分子名称: | 1-[4-[6-chloranyl-7-(5-methyl-1~{H}-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Phillips, C. | | 登録日 | 2019-10-17 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
6O6L
 
 | | The Structure of EgtB(Cabther) in complex with Hercynine | | 分子名称: | EgtB (Cabther), FE (III) ION, N,N,N-trimethyl-histidine | | 著者 | Irani, S, Zhang, Y. | | 登録日 | 2019-03-07 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
7XWX
 
 | | Crystal structure of SARS-CoV-2 N-CTD | | 分子名称: | Nucleoprotein, PHOSPHATE ION | | 著者 | Luan, X.D, Li, X.M, Li, Y.F. | | 登録日 | 2022-05-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
 | | Crystal structure of SARS-CoV-2 N-NTD | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | 著者 | Luan, X.D, Li, X.M, Li, Y.F. | | 登録日 | 2022-05-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XWZ
 
 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | 著者 | Luan, X.D, Li, X.M, Li, Y.F. | | 登録日 | 2022-05-27 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
6O6M
 
 | | The Structure of EgtB (Cabther) | | 分子名称: | EgtB (Cabther), FE (III) ION, GLYCEROL | | 著者 | Irani, S, Zhang, Y. | | 登録日 | 2019-03-07 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.506 Å) | | 主引用文献 | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
8GSF
 
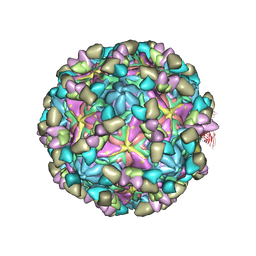 | |
8GSD
 
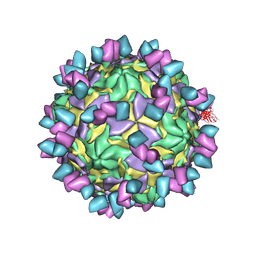 | | Echovirus3 full particle in complex with 6D10 Fab | | 分子名称: | Genome polyprotein, Genome polyprotein (Fragment), Heavy chain of 6D10, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-09-06 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
8GSE
 
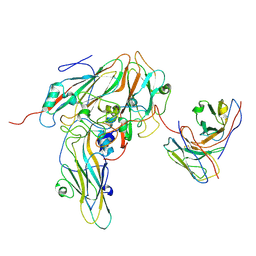 | |
8GSC
 
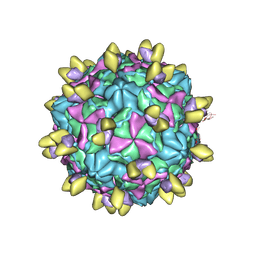 | | Echovirus3 A-particle in complex with 6D10 Fab | | 分子名称: | Heavy chain of 6D10, Light chain of 6D10, VP1, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-09-06 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural Basis for the Immunogenicity of the C-Terminus of VP1 of Echovirus 3 Revealed by the Binding of a Neutralizing Antibody.
Viruses, 14, 2022
|
|
7X6S
 
 | |
