6E9Z
 
 | | DHF119 filament | | 分子名称: | DHF119 filament | | 著者 | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | 登録日 | 2018-08-01 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6E9V
 
 | | DHF79 filament | | 分子名称: | DHF79 filament | | 著者 | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | 登録日 | 2018-08-01 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
8CTO
 
 | |
8CUN
 
 | |
8CWA
 
 | |
6TMS
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | 分子名称: | SULFATE ION, a novel designed pore protein, affinity purification tag | | 著者 | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | 登録日 | 2019-12-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
6TJ1
 
 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | 分子名称: | De novo designed WSHC6, purification tag | | 著者 | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | 登録日 | 2019-11-23 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
7N1J
 
 | |
7N1K
 
 | |
7N3T
 
 | | TrkA ECD complex with designed miniprotein ligand | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Jude, K.M, Cao, L, Garcia, K.C. | | 登録日 | 2021-06-01 | | 公開日 | 2022-04-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Design of protein-binding proteins from the target structure alone.
Nature, 605, 2022
|
|
3L56
 
 | | Crystal structure of the large c-terminal domain of polymerase basic protein 2 from influenza virus a/viet nam/1203/2004 (h5n1) | | 分子名称: | Polymerase PB2 | | 著者 | Staker, B.L, Edwards, T, Eric, S, Raymond, A, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2009-12-21 | | 公開日 | 2010-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Biological and structural characterization of a host-adapting amino acid in influenza virus.
Plos Pathog., 6, 2010
|
|
6DG6
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic | | 分子名称: | Neoleukin-2/15 | | 著者 | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | 登録日 | 2018-05-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
6DG5
 
 | | Structure of a de novo designed Interleukin-2/Interleukin-15 mimetic complex with IL-2Rb and IL-2Rg | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | 著者 | Jude, K.M, Silva, D.-A, Yu, S, Baker, D, Garcia, K.C. | | 登録日 | 2018-05-16 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.516 Å) | | 主引用文献 | De novo design of potent and selective mimics of IL-2 and IL-15.
Nature, 565, 2019
|
|
8GJI
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | 分子名称: | GCG binder, Glucagon | | 著者 | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | 登録日 | 2023-03-15 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GJG
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | 分子名称: | gluc_A04_0005, gluc_A04_0005 Binder | | 著者 | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | 登録日 | 2023-03-15 | | 公開日 | 2024-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8GLT
 
 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | 分子名称: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | 著者 | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | 登録日 | 2023-03-23 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
6WRW
 
 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | 著者 | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRX
 
 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRV
 
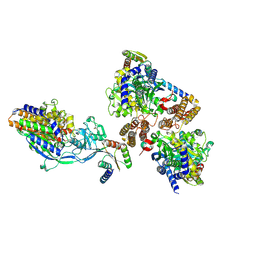 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | 著者 | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8FRE
 
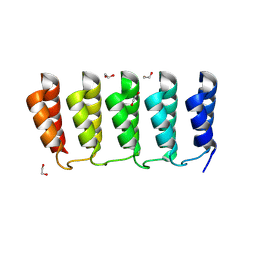 | | Designed loop protein RBL4 | | 分子名称: | 1,2-ETHANEDIOL, RBL4 | | 著者 | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | 登録日 | 2023-01-06 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
8FRF
 
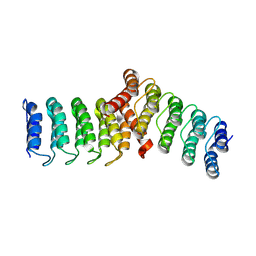 | | Homodimeric designed loop protein RBL7_C2_3 | | 分子名称: | MALONATE ION, RBL7_C2_3 | | 著者 | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | 登録日 | 2023-01-06 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
5KOB
 
 | |
7UHB
 
 | |
7UHC
 
 | |
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | 分子名称: | TMH4C4 | | 著者 | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | 登録日 | 2020-03-16 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
