7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | 著者 | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | 登録日 | 2020-11-05 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7CMZ
 
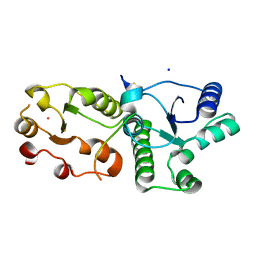 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | 分子名称: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | 著者 | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | 登録日 | 2020-07-29 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
7FAH
 
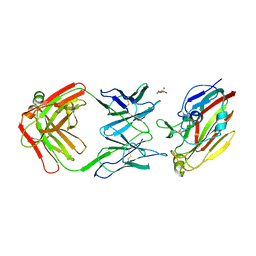 | | Immune complex of head region of CA09 HA and neutralizing antibody 12H5 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | Li, T.T, Xue, W.H, Gu, Y, Li, S.W. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.151 Å) | | 主引用文献 | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
8GPJ
 
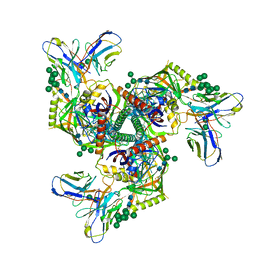 | | HIV-1 Env X16 UFO in complex with 8ANC195 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8ANC195 Fab heavy chain, ... | | 著者 | Niu, J, Xu, Y.W, Yang, B. | | 登録日 | 2022-08-26 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs.
Nat Commun, 14, 2023
|
|
8GPG
 
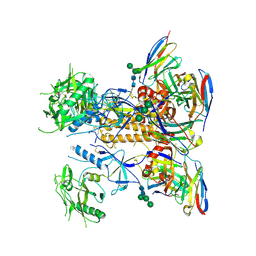 | | HIV-1 Env X18 UFO in complex with F6 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, F6 Fab heavy chain, ... | | 著者 | Niu, J, Xu, Y.W, Yang, B. | | 登録日 | 2022-08-26 | | 公開日 | 2023-08-09 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structures and immune recognition of Env trimers from two Asia prevalent HIV-1 CRFs.
Nat Commun, 14, 2023
|
|
5ZUV
 
 | | Crystal Structure of the Human Coronavirus 229E HR1 motif in complex with pan-CoVs inhibitor EK1 | | 分子名称: | CHLORIDE ION, Spike glycoprotein,Spike glycoprotein,inhibitor EK1 | | 著者 | Yan, L, Yang, B, Wilson, I.A. | | 登録日 | 2018-05-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike.
Sci Adv, 5, 2019
|
|
5ZVM
 
 | |
5ZVK
 
 | |
6A3V
 
 | | Complex structure of human 4-1BB and 4-1BBL | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | 著者 | Li, Y, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | 登録日 | 2018-06-17 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.391 Å) | | 主引用文献 | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
6AHU
 
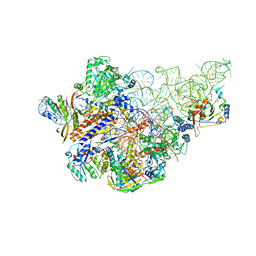 | | Cryo-EM structure of human Ribonuclease P with mature tRNA | | 分子名称: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | 著者 | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | 登録日 | 2018-08-20 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6A3W
 
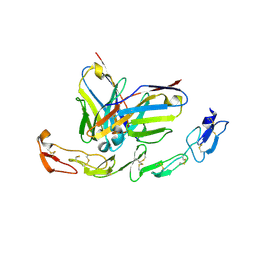 | | Complex structure of 4-1BB and utomilumab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 9, utomilumab VH, ... | | 著者 | Li, Y, Tan, S, Zhang, C, Chai, Y, Qi, J, Tien, P, Gao, S, Gao, G.F. | | 登録日 | 2018-06-17 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Limited Cross-Linking of 4-1BB by 4-1BB Ligand and the Agonist Monoclonal Antibody Utomilumab.
Cell Rep, 25, 2018
|
|
6ABO
 
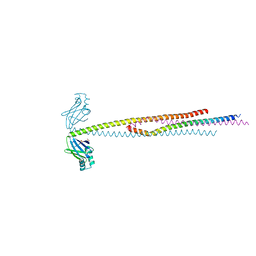 | | human XRCC4 and IFFO1 complex | | 分子名称: | DNA repair protein XRCC4, GLYCEROL, Intermediate filament family orphan 1, ... | | 著者 | Li, J, Liu, L, Liang, H, Liu, Y, Xu, D. | | 登録日 | 2018-07-23 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Nat.Cell Biol., 21, 2019
|
|
6AHV
 
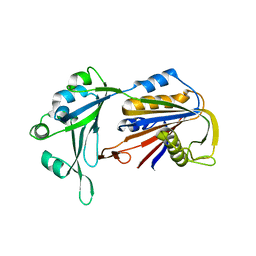 | | Crystal structure of human RPP40 | | 分子名称: | Ribonuclease P protein subunit p40 | | 著者 | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | 登録日 | 2018-08-20 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
6AHR
 
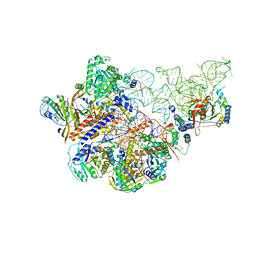 | | Cryo-EM structure of human Ribonuclease P | | 分子名称: | H1 RNA, Ribonuclease P protein subunit p14, Ribonuclease P protein subunit p20, ... | | 著者 | Wu, J, Niu, S, Tan, M, Lan, P, Lei, M. | | 登録日 | 2018-08-20 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.92 Å) | | 主引用文献 | Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Cell, 175, 2018
|
|
7DL9
 
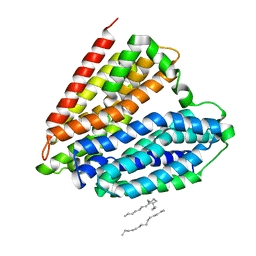 | |
7DLA
 
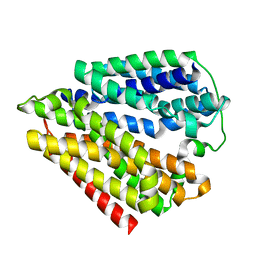 | |
6CSJ
 
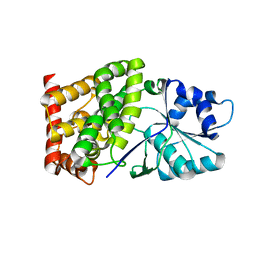 | |
7DRV
 
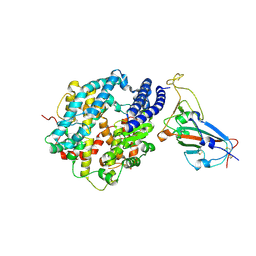 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | 登録日 | 2020-12-29 | | 公開日 | 2021-08-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.09 Å) | | 主引用文献 | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
6L8O
 
 | |
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | 分子名称: | DNA repair protein RAD5, ZINC ION | | 著者 | Shen, M, Xiang, S. | | 登録日 | 2019-11-06 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8R
 
 | | membrane-bound PD-L1-CD | | 分子名称: | Programmed cell death 1 ligand 1 | | 著者 | Maorong, W, Cao, Y, Bin, W, Bo, O. | | 登録日 | 2019-11-07 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain.
Nat Commun, 12, 2021
|
|
6LS4
 
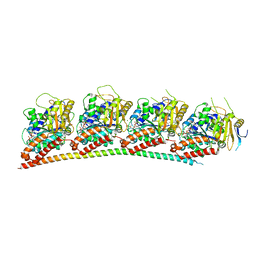 | | A novel anti-tumor agent S-40 in complex with tubulin | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(4-cyclopropylphenyl)sulfonylamino]-4-methyl-N-(pyridin-3-ylmethyl)benzamide, GLYCEROL, ... | | 著者 | Du, T, Lin, S, Ji, M, Xue, N, Liu, Y, Zhang, K, Lu, D, Chen, X, Xu, H. | | 登録日 | 2020-01-17 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A novel orally active microtubule destabilizing agent S-40 targets the colchicine-binding site and shows potent antitumor activity.
Cancer Lett., 495, 2020
|
|
7DFG
 
 | |
7DFH
 
 | |
8WHA
 
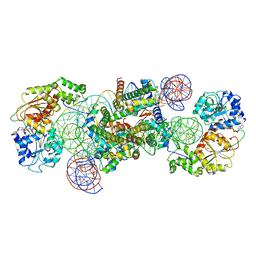 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
