5E1O
 
 | | Crystal structure of NTMT1 in complex with RPKRIA peptide | | 分子名称: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | 著者 | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2015-09-29 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E1B
 
 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | 分子名称: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | 著者 | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2015-09-29 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5VAR
 
 | |
5V9L
 
 | |
5TEF
 
 | | Crystal structure of Gemin5 WD40 repeats in complex with m7GpppG | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, GLYCEROL, Gem-associated protein 5, ... | | 著者 | Chao, X, Tempel, W, Bian, C, He, H, Cerovina, T, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-09-21 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | 分子名称: | Calmodulin-1, Caspase-4, OspC3, ... | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5V71
 
 | | KRAS G12C in bound to quinazoline based switch II pocket (SWIIP) binder | | 分子名称: | 1-{4-[6-chloro-7-(2-fluorophenyl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Westover, K, Lu, J. | | 登録日 | 2017-03-17 | | 公開日 | 2017-08-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.228 Å) | | 主引用文献 | KRAS G12C Drug Development: Discrimination between Switch II Pocket Configurations Using Hydrogen/Deuterium-Exchange Mass Spectrometry.
Structure, 25, 2017
|
|
5V9O
 
 | | KRAS G12C inhibitor | | 分子名称: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Westover, K, Lu, J. | | 登録日 | 2017-03-23 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Potent and Selective Covalent Quinazoline Inhibitors of KRAS G12C.
Cell Chem Biol, 24, 2017
|
|
7WZS
 
 | |
7WR4
 
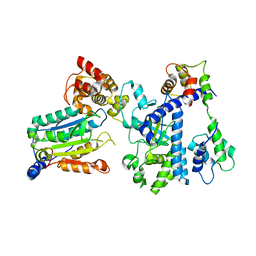 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | 分子名称: | Calmodulin-1, Caspase-4, OspC3 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
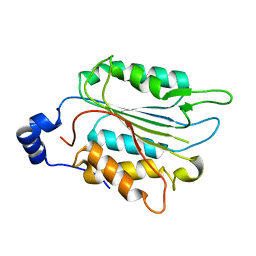 | | P32 of caspase-4 C258A mutant | | 分子名称: | Caspase-4 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
7WR6
 
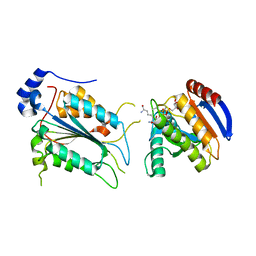 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | 分子名称: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | 分子名称: | Caspase-4, OspC3 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | 分子名称: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6L30
 
 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | 分子名称: | Protein ECT2 | | 著者 | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | 登録日 | 2019-10-07 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6L54
 
 | | Structure of SMG189 | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein SMG8, ... | | 著者 | Xu, Y, Qi, Y. | | 登録日 | 2019-10-22 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.43 Å) | | 主引用文献 | Cryo-EM structure of SMG1-SMG8-SMG9 complex.
Cell Res., 29, 2019
|
|
2P4S
 
 | | Structure of Purine Nucleoside Phosphorylase from Anopheles gambiae in complex with DADMe-ImmH | | 分子名称: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | 著者 | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | 登録日 | 2007-03-13 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Anopheles gambiae purine nucleoside phosphorylase: catalysis, structure, and inhibition.
Biochemistry, 46, 2007
|
|
6L53
 
 | | Structure of SMG1 | | 分子名称: | Serine/threonine-protein kinase SMG1 | | 著者 | Xu, Y, Qi, Y. | | 登録日 | 2019-10-22 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Cryo-EM structure of SMG1-SMG8-SMG9 complex.
Cell Res., 29, 2019
|
|
2PER
 
 | |
3BWN
 
 | | L-tryptophan aminotransferase | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | 著者 | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | 登録日 | 2008-01-10 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
3GQA
 
 | |
3GQK
 
 | |
3GQ9
 
 | |
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Chen, S.J, Ye, F. | | 登録日 | 2016-08-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
