7L0O
 
 | |
4FEA
 
 | |
5TUY
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor MS0124 | | 分子名称: | 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-(morpholin-4-yl)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | 著者 | Babault, N, Xiong, Y, Liu, J, Jin, J. | | 登録日 | 2016-11-07 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5VSC
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 13 | | 分子名称: | 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | 著者 | Babault, N, Xiong, Y, Liu, J, Jin, J. | | 登録日 | 2017-05-11 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
6X3P
 
 | | Co-structure of BTK kinase domain with L-005298385 inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{8-amino-3-[(6R,8aS)-3-oxooctahydroindolizin-6-yl]imidazo[1,5-a]pyrazin-1-yl}-3-(cyclopropyloxy)-N-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | 著者 | Fischmann, T.O. | | 登録日 | 2020-05-21 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | Potent, non-covalent reversible BTK inhibitors with 8-amino-imidazo[1,5-a]pyrazine core featuring 3-position bicyclic ring substitutes.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | 著者 | Babault, N, Xiong, Y, Liu, J, Jin, J. | | 登録日 | 2017-05-11 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSE
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 17: N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine | | 分子名称: | Histone-lysine N-methyltransferase EHMT2, N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | 著者 | Babault, N, Xiong, Y, Liu, J, Jin, J. | | 登録日 | 2017-05-11 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSD
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 13 | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, GLYCEROL, ... | | 著者 | Babault, N, Xiong, Y, Liu, J, Jin, J. | | 登録日 | 2017-05-11 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
2FZ0
 
 | | Identification of yeast R-SNARE Nyv1p as a novel longin domain protein | | 分子名称: | v-SNARE component of the vacuolar SNARE complex involved in vesicle fusion; inhibits ATP-dependent Ca(2+) transport activity of Pmc1p in the vacuolar membrane; Nyv1p | | 著者 | Wen, W, Zhang, M. | | 登録日 | 2006-02-08 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Identification of the Yeast R-SNARE Nyv1p as a Novel Longin Domain-containing Protein
Mol.Cell.Biol., 17, 2006
|
|
4D2K
 
 | |
4DPV
 
 | | PARVOVIRUS/DNA COMPLEX | | 分子名称: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | 著者 | Chapman, M.S, Rossmann, M.G. | | 登録日 | 1996-02-01 | | 公開日 | 1997-04-01 | | 最終更新日 | 2023-04-19 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | 分子名称: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | 著者 | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | 分子名称: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | 著者 | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
6JBT
 
 | | Complex structure of toripalimab-Fab and PD-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | 著者 | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | 登録日 | 2019-01-26 | | 公開日 | 2019-06-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
4EJN
 
 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | 分子名称: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | 著者 | Eathiraj, S. | | 登録日 | 2012-04-06 | | 公開日 | 2012-05-23 | | 最終更新日 | 2013-01-02 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
8FG8
 
 | | Catalytic domain of GtfB in complex with inhibitor 2-[(2,4,5-Trihydroxyphenyl)methylidene]-1-benzofuran-3-one | | 分子名称: | (2Z)-2-[(2,4,5-trihydroxyphenyl)methylidene]-1-benzofuran-3(2H)-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | 著者 | Schormann, N, Deivanayagam, C, Velu, S. | | 登録日 | 2022-12-12 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Hydrogel-Encapsulated Biofilm Inhibitors Abrogate the Cariogenic Activity of Streptococcus mutans .
J.Med.Chem., 66, 2023
|
|
6D74
 
 | |
5HVW
 
 | | Monomeric IgG4 Fc | | 分子名称: | GLYCEROL, Ig gamma-4 chain C region, ZINC ION, ... | | 著者 | Oganesyan, V.Y, Shan, L, Dall'Acqua, W.F. | | 登録日 | 2016-01-28 | | 公開日 | 2016-08-17 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Generation and Characterization of an IgG4 Monomeric Fc Platform.
Plos One, 11, 2016
|
|
5J87
 
 | |
3KXL
 
 | | crystal structure of SsGBP mutation variant G235S | | 分子名称: | GTP-binding protein (HflX), THIOCYANATE ION | | 著者 | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | 登録日 | 2009-12-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KXI
 
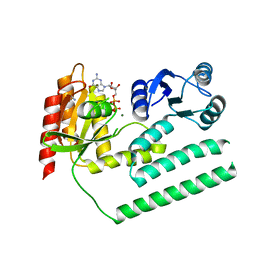 | | crystal structure of SsGBP and GDP complex | | 分子名称: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | 登録日 | 2009-12-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
5YGI
 
 | |
5IN1
 
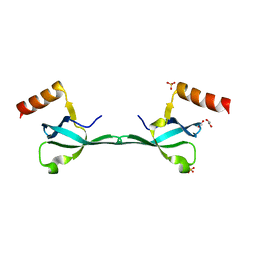 | | Crystal Structure of the MRG701 chromodomain | | 分子名称: | 1,2-ETHANEDIOL, MRG701, SULFATE ION | | 著者 | Huang, Y, Liu, Y. | | 登録日 | 2016-03-07 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants
Protein Cell, 7, 2016
|
|
3I6G
 
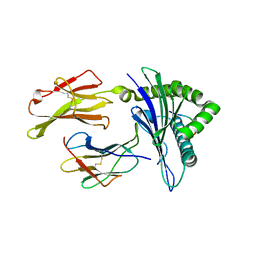 | | Newly identified epitope Mn2 from SARS-CoV M protein complexed withHLA-A*0201 | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | 著者 | Liu, J. | | 登録日 | 2009-07-07 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes.
J.INFECT.DIS., 202, 2010
|
|
5KCV
 
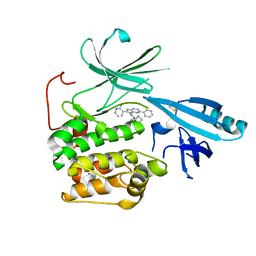 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | 分子名称: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | 著者 | Eathiraj, S. | | 登録日 | 2016-06-07 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
