7FBD
 
 | | De novo design protein D53 with MBP tag | | 分子名称: | Maltodextrin-binding protein,De novo design protein D53 | | 著者 | Bin, H. | | 登録日 | 2021-07-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7FBB
 
 | | De novo design protein D12 with MBP tag | | 分子名称: | Maltodextrin-binding protein,de novo designed protein D12 | | 著者 | Bin, H. | | 登録日 | 2021-07-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.307 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6W8D
 
 | | Structure of DNMT3A (R882H) in complex with CGT DNA | | 分子名称: | CGT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | 著者 | Anteneh, H, Song, J. | | 登録日 | 2020-03-20 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.598 Å) | | 主引用文献 | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
7FBC
 
 | | De novo design protein D22 with MBP tag | | 分子名称: | Maltodextrin-binding protein,De novo design protein D22 | | 著者 | Bin, H. | | 登録日 | 2021-07-09 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
6BSH
 
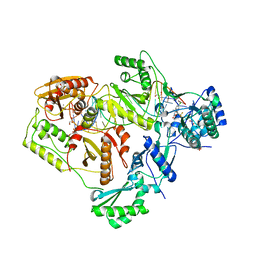 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in the RNA hydrolysis mode | | 分子名称: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DNA (5'-D(*GP*TP*AP*TP*GP*CP*CP*AP*CP*TP*AP*GP*TP*TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3'), ... | | 著者 | Tian, L, Kim, M, Yang, W. | | 登録日 | 2017-12-03 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.649 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7VZR
 
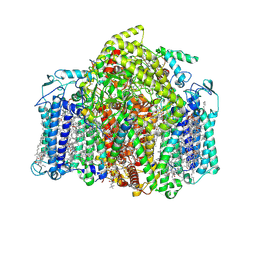 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | 分子名称: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | 著者 | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | 登録日 | 2021-11-16 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.22 Å) | | 主引用文献 | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
6PSO
 
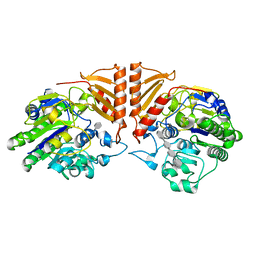 | | Crystal structure of PsS1_19B C77S in complex with iota-neocarratetraose | | 分子名称: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Hettle, A.G, Boraston, A.B. | | 登録日 | 2019-07-13 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6PT6
 
 | |
7VZG
 
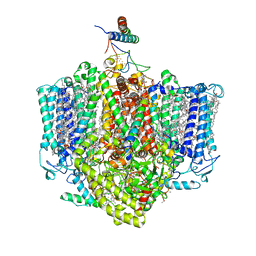 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | 分子名称: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | 著者 | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | 登録日 | 2021-11-16 | | 公開日 | 2023-02-22 | | 実験手法 | ELECTRON MICROSCOPY (2.61 Å) | | 主引用文献 | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
3WMC
 
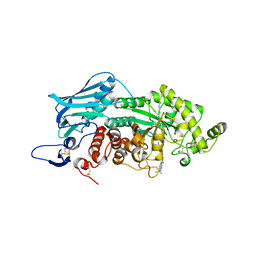 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(dimethylamino)-2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, Beta-hexosaminidase | | 著者 | Chen, L, Zhou, Y, Chen, L, Yang, Q. | | 登録日 | 2013-11-16 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.095 Å) | | 主引用文献 | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
4YQH
 
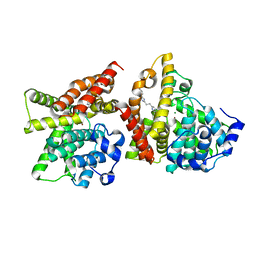 | | 2-[2-(4-Phenyl-1H-imidazol-2-yl)ethyl]quinoxaline (Sunovion Compound 14) co-crystallized with PDE10A | | 分子名称: | 2-[2-(4-phenyl-1H-imidazol-2-yl)ethyl]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Burdi, D, Herman, L, Wang, T. | | 登録日 | 2015-03-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.308 Å) | | 主引用文献 | Evolution and synthesis of novel orally bioavailable inhibitors of PDE10A.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4YS7
 
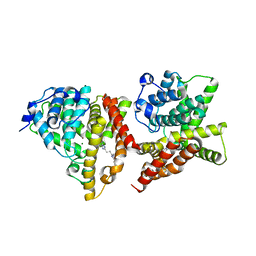 | | Co-crystal structure of 2-[2-(5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl)ethyl]-3-methyl-3H-imidazo[4,5-f]quinoline (compound 39) with PDE10A | | 分子名称: | 2-[2-(5,8-dimethyl[1,2,4]triazolo[1,5-a]pyrazin-2-yl)ethyl]-3-methyl-3H-imidazo[4,5-f]quinoline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Burdi, D.F, Herman, L, Wang, T. | | 登録日 | 2015-03-16 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Evolution and synthesis of novel orally bioavailable inhibitors of PDE10A.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8UVX
 
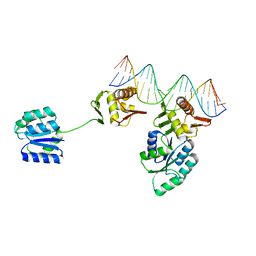 | | CosR DNA bound form I | | 分子名称: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | 著者 | Zhang, Z. | | 登録日 | 2023-11-05 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
3EE1
 
 | |
5XJZ
 
 | | Structure of DNA1-Ag complex | | 分子名称: | DNA (5'-D(*GP*CP*AP*CP*GP*CP*GP*C)-3'), SILVER ION, SPERMINE | | 著者 | Liu, H.H, Gan, J.H. | | 登録日 | 2017-05-04 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XNE
 
 | |
3SWJ
 
 | | Crystal structure of Campylobacter jejuni ChuZ | | 分子名称: | AZIDE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein | | 著者 | Hu, Y. | | 登録日 | 2011-07-14 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.409 Å) | | 主引用文献 | Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode.
Biochem.Biophys.Res.Commun., 415, 2011
|
|
8ECG
 
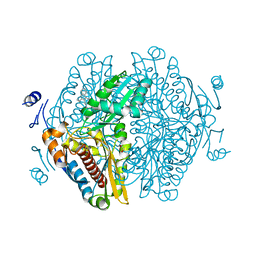 | | Complex of HMG1 with pitavastatin | | 分子名称: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase 1, GLYCEROL, Pitavastatin | | 著者 | Haywood, J, Bond, C.S. | | 登録日 | 2022-09-01 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | A fungal tolerance trait and selective inhibitors proffer HMG-CoA reductase as a herbicide mode-of-action.
Nat Commun, 13, 2022
|
|
4P6A
 
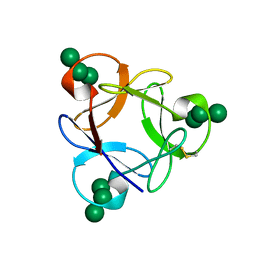 | | Crystal structure of a potent anti-HIV lectin actinohivin in complex with alpha-1,2-mannotriose | | 分子名称: | Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | 著者 | Zhang, F, Hoque, M.M, Suzuki, K, Tsunoda, M, Naomi, O, Tanaka, H, Takenaka, A. | | 登録日 | 2014-03-23 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | The characteristic structure of anti-HIV actinohivin in complex with three HMTG D1 chains of HIV-gp120.
Chembiochem, 15, 2014
|
|
7CSX
 
 | |
3C4E
 
 | |
5NFS
 
 | |
4M37
 
 | |
5WUS
 
 | |
6C0F
 
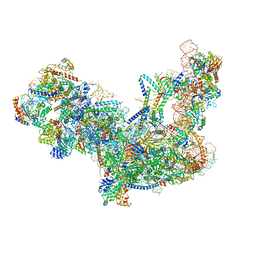 | | Yeast nucleolar pre-60S ribosomal subunit (state 2) | | 分子名称: | 5.8S rRNA, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Sanghai, Z.A, Miller, L, Barandun, J, Hunziker, M, Chaker-Margot, M, Klinge, S. | | 登録日 | 2017-12-29 | | 公開日 | 2018-03-14 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Modular assembly of the nucleolar pre-60S ribosomal subunit.
Nature, 556, 2018
|
|
