8PJN
 
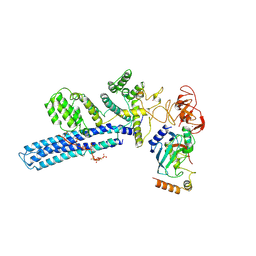 | | Catalytic module of human CTLH E3 ligase bound to multiphosphorylated UBE2H~ubiquitin | | 分子名称: | E3 ubiquitin-protein transferase MAEA, E3 ubiquitin-protein transferase RMND5A, Ubiquitin, ... | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, R.J, Schulman, B.A. | | 登録日 | 2023-06-23 | | 公開日 | 2024-01-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Multisite phosphorylation dictates selective E2-E3 pairing as revealed by Ubc8/UBE2H-GID/CTLH assemblies.
Mol.Cell, 84, 2024
|
|
8PD0
 
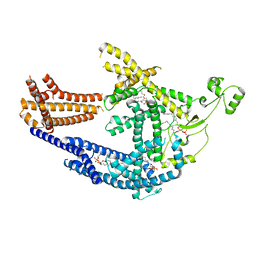 | | cryo-EM structure of Doa10 in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-11 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
8PDA
 
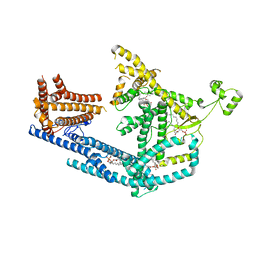 | | cryo-EM structure of Doa10 with RING domain in MSP1E3D1 | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ERAD-associated E3 ubiquitin-protein ligase DOA10 | | 著者 | Botsch, J.J, Braeuning, B, Schulman, B.A. | | 登録日 | 2023-06-12 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Doa10/MARCH6 architecture interconnects E3 ligase activity with lipid-binding transmembrane channel to regulate SQLE.
Nat Commun, 15, 2024
|
|
5L9U
 
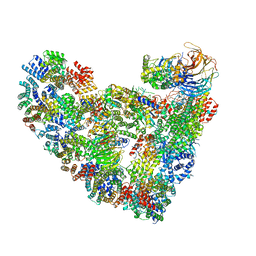 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | 登録日 | 2016-06-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
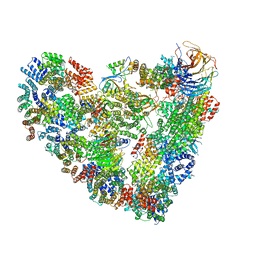 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | 登録日 | 2016-06-11 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
4RG6
 
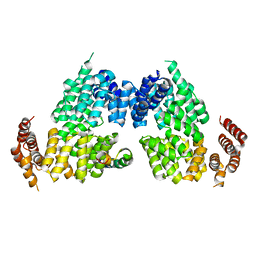 | | Crystal structure of APC3-APC16 complex | | 分子名称: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | 著者 | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | 登録日 | 2014-09-29 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4RG9
 
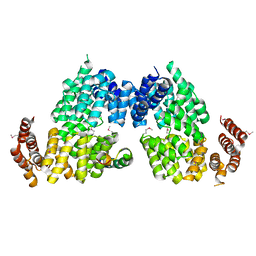 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | 分子名称: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | 著者 | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | 登録日 | 2014-09-29 | | 公開日 | 2014-12-24 | | 最終更新日 | 2017-08-02 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
7ONI
 
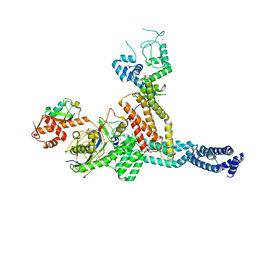 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | 分子名称: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | 著者 | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | 登録日 | 2021-05-25 | | 公開日 | 2021-09-15 | | 最終更新日 | 2021-10-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
7Z8B
 
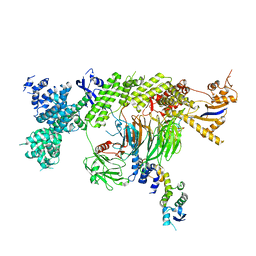 | |
7ZBZ
 
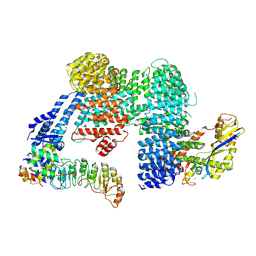 | |
7Z8R
 
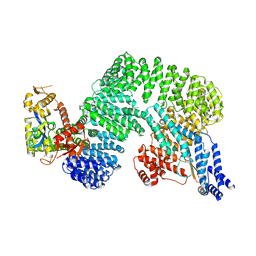 | | CAND1-CUL1-RBX1 | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Schulman, B.A. | | 登録日 | 2022-03-18 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8T
 
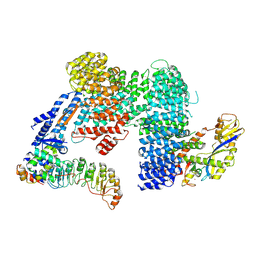 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Schulman, B.A. | | 登録日 | 2022-03-18 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8V
 
 | |
7ZBW
 
 | | CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged | | 分子名称: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Schulman, B.A. | | 登録日 | 2022-03-24 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | 分子名称: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-10-27 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | 分子名称: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | 著者 | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | 登録日 | 2017-10-27 | | 公開日 | 2018-09-26 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6WY6
 
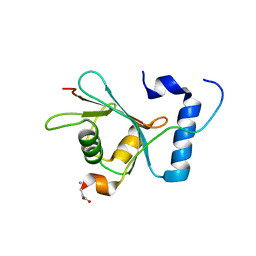 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | 分子名称: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | 著者 | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | 登録日 | 2020-05-12 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
6SWY
 
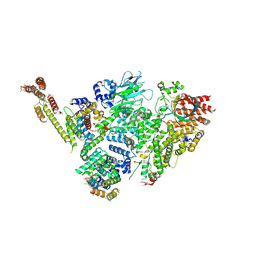 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | 分子名称: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | 著者 | Qiao, S, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-09-24 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
6TTU
 
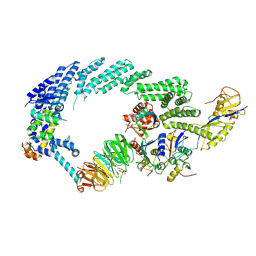 | | Ubiquitin Ligation to substrate by a cullin-RING E3 ligase at 3.7A resolution: NEDD8-CUL1-RBX1 N98R-SKP1-monomeric b-TRCP1dD-IkBa-UB~UBE2D2 | | 分子名称: | CYS-LYS-LYS-ALA-ARG-HIS-ASP-SEP-GLY, Cullin-1, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Baek, K, Prabu, J.R, Schulman, B.A. | | 登録日 | 2019-12-30 | | 公開日 | 2020-02-12 | | 最終更新日 | 2020-03-04 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly.
Nature, 578, 2020
|
|
8EBM
 
 | |
8EBN
 
 | |
8EBL
 
 | |
7NS3
 
 | | Substrate receptor scaffolding module of yeast Chelator-GID SR4 E3 ubiquitin ligase bound to Fbp1 substrate | | 分子名称: | BJ4_G0018240.mRNA.1.CDS.1, Fructose-bisphosphatase, Glucose-induced degradation protein 8, ... | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-07-07 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NSB
 
 | | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | 分子名称: | Glucose-induced degradation protein 7, Glucose-induced degradation protein 8, Vacuolar import and degradation protein 30 | | 著者 | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NS5
 
 | | Structure of yeast Fbp1 (Fructose-1,6-bisphosphatase 1) | | 分子名称: | Fructose-1,6-bisphosphatase, MAGNESIUM ION, PHOSPHATE ION | | 著者 | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | 登録日 | 2021-03-05 | | 公開日 | 2021-05-05 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
