7PTJ
 
 | |
5FQM
 
 | | Last common ancestor of Gram Negative Bacteria (GNCA) Class A beta- lactamase | | 分子名称: | GLYCEROL, GNCA BETA LACTAMASE, SULFATE ION | | 著者 | Martinez Rodriguez, S, Gavira, J.A, Risso, V.A, Sanchez Ruiz, J.M. | | 登録日 | 2015-12-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
6G5Z
 
 | |
6G60
 
 | |
5FQJ
 
 | | W229D mutant of the last common ancestor of Gram-negative bacteria (GNCA) beta-lactamase bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, GNCA LACTAMASE W229D | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.271 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5FQQ
 
 | | Last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GNCA4 LACTAMASE | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Risso, V.A, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-14 | | 公開日 | 2016-12-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5G3O
 
 | | Bacillus cereus formamidase (BceAmiF) inhibited with urea. | | 分子名称: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, FORMAMIDASE, ... | | 著者 | Gavira, J.A, Martinez-Rodriguez, S, Conejero-Muriel, M. | | 登録日 | 2016-04-29 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
5FQK
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A bound to 5(6)-nitrobenzotriazole (TS-analog) | | 分子名称: | 6-NITROBENZOTRIAZOLE, GNCA4 LACTAMASE W229D AND F290W | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.767 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
5G3P
 
 | | Bacillus cereus formamidase (BceAmiF) acetylated at the active site. | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gavira, J.A, Conejero-Muriel, M, Martinez-Rodriguez, S. | | 登録日 | 2016-04-29 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
5FQI
 
 | | W229D and F290W mutant of the last common ancestor of Gram-negative bacteria (GNCA4) beta-lactamase class A | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gavira, J.A, Risso, V.A, Martinez-Rodriguez, S, Sanchez-Ruiz, J.M. | | 登録日 | 2015-12-11 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | De novo active sites for resurrected Precambrian enzymes.
Nat Commun, 8, 2017
|
|
3VCY
 
 | | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase), from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin. | | 分子名称: | GLYCEROL, PHOSPHATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | 著者 | Bensen, D.C, Rodriguez, S, Nix, J, Cunningham, M.L, Tari, L.W. | | 登録日 | 2012-01-04 | | 公開日 | 2012-04-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.925 Å) | | 主引用文献 | Structure of MurA (UDP-N-acetylglucosamine enolpyruvyl transferase) from Vibrio fischeri in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
8BGA
 
 | | Structure of Mpro in complex with FGA146 | | 分子名称: | 3C-like proteinase nsp5, 4-methoxy-~{N}-[(2~{S})-4-methyl-1-[[(2~{S})-4-nitro-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide | | 著者 | Medrano, F.J, Romero, A. | | 登録日 | 2022-10-27 | | 公開日 | 2023-11-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.982 Å) | | 主引用文献 | Peptidyl nitroalkene inhibitors of main protease rationalized by computational and crystallographic investigations as antivirals against SARS-CoV-2.
Commun Chem, 7, 2024
|
|
4JZ4
 
 | | Crystal structure of chicken c-Src-SH3 domain: monomeric form | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Camara-Artigas, A. | | 登録日 | 2013-04-02 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
4JZ3
 
 | |
2Z70
 
 | |
4K4O
 
 | | The DNA Gyrase B ATP binding domain of Enterococcus faecalis in complex with a small molecule inhibitor | | 分子名称: | 6-fluoro-4-[(3aR,6aR)-hexahydropyrrolo[3,4-b]pyrrol-5(1H)-yl]-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Lam, T, Tari, L.W. | | 登録日 | 2013-04-12 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A new class of type IIA topoisomerase inhibitors with broad-spectrum antibacterial
activity
To be Published
|
|
4KSG
 
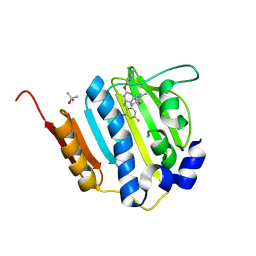 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (4-[(1S,5R,6R)-6-AMINO-1-METHYL-3-AZABICYCLO[3.2.0]HEPT-3-YL]-6-FLUORO-N-METHYL-2-[(2-METHYLPYRIMIDIN-5-YL)OXY]-9H-PYRIMIDO[4,5-B]INDOL-8-AMINE) | | 分子名称: | 4-[(1S,5R,6R)-6-amino-1-methyl-3-azabicyclo[3.2.0]hept-3-yl]-6-fluoro-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | 登録日 | 2013-05-17 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
4KTN
 
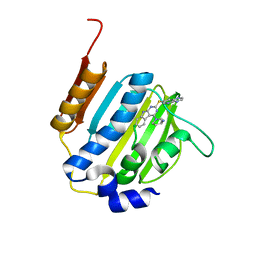 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor ((3S)-1-[2-(PYRIDO[2,3-B]PYRAZIN-7-YLSULFANYL)-9H-PYRIMIDO[4,5-B]INDOL-4-YL]PYRROLIDIN-3-AMINE) | | 分子名称: | (3S)-1-[2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-9H-pyrimido[4,5-b]indol-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B | | 著者 | Bensen, D.C, Akers-rodriguez, S, Tari, L.W. | | 登録日 | 2013-05-20 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
4KSH
 
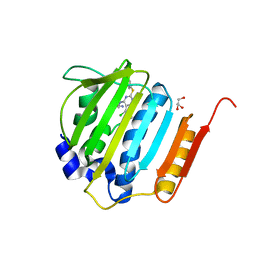 | | Dna gyrase atp binding domain of enterococcus faecalis in complex with a small molecule inhibitor (7-({4-[(3R)-3-AMINOPYRROLIDIN-1-YL]-5-CHLORO-6-ETHYL-7H-PYRROLO[2,3-D]PYRIMIDIN-2-YL}SULFANYL)-1,5-NAPHTHYRIDIN-1(4H)-OL) | | 分子名称: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1,5-naphthyridin-1(4H)-ol, DNA gyrase subunit B, GLYCEROL | | 著者 | Bensen, D.C, Akers-Rodriguez, S, Tari, L.W. | | 登録日 | 2013-05-17 | | 公開日 | 2014-01-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A new class of type iia topoisomerase inhibitors with broad-spectrum antibacterial activity
To be Published
|
|
3DWT
 
 | |
6HW1
 
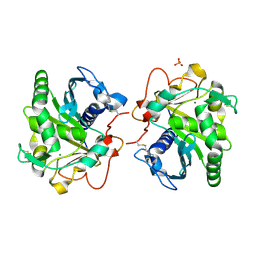 | | ROOM TEMPERATURE STRUCTURE OF LIPASE FROM T. LANUGINOSA AT 2.5 A RESOLUTION IN CHIPX MICROFLUIDIC DEVICE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase, MAGNESIUM ION, ... | | 著者 | Gavira, J.A, Fernadez-Penas, R, Martinez-Rodriguez, S, Verdugo-Escamilla, C. | | 登録日 | 2018-10-11 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
4OMO
 
 | |
3S8J
 
 | |
3S8K
 
 | |
6GCV
 
 | | Ligand binding domain (LBD) of the p. aeruginosa nitrate receptor McpN | | 分子名称: | ACETATE ION, Chemotaxis transducer, NITRATE ION, ... | | 著者 | Gavira, J.A, Krell, T, Martin-Mora, D, Ortega, A. | | 登録日 | 2018-04-19 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | The Molecular Mechanism of Nitrate Chemotaxis via Direct Ligand Binding to the PilJ Domain of McpN.
MBio, 10, 2019
|
|
