1OXG
 
 | | Crystal structure of a complex formed between organic solvent treated bovine alpha-chymotrypsin and its autocatalytically produced highly potent 14-residue peptide at 2.2 resolution | | 分子名称: | Chymotrypsinogen A, SULFATE ION | | 著者 | Singh, N, Jabeen, T, Sharma, S, Roy, I, Gupta, M.N, Bilgrami, S, Singh, T.P. | | 登録日 | 2003-04-02 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Detection of native peptides as potent inhibitors of enzymes. Crystal structure of the complex formed between treated bovine alpha-chymotrypsin and an autocatalytically produced fragment, IIe-Val-Asn-Gly-Glu-Glu-Ala-Val-Pro-Gly-Ser-Trp-Pro-Trp, at 2.2 angstroms resolution.
Febs J., 272, 2005
|
|
4OOM
 
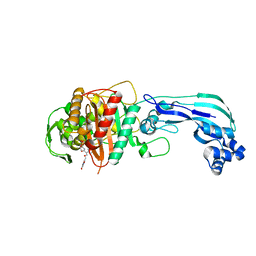 | | Crystal structure of PBP3 in complex with BAL30072 ((2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide) | | 分子名称: | (2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide, Cell division protein FtsI [Peptidoglycan synthetase] | | 著者 | Han, S, Caspers, N, Knafels, J.D. | | 登録日 | 2014-02-03 | | 公開日 | 2014-05-07 | | 最終更新日 | 2014-05-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
3EKA
 
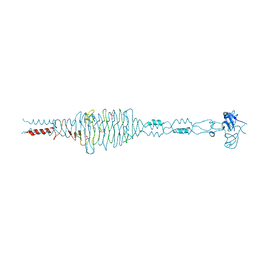 | | Crystal structure of the complex of hyaluranidase trimer with ascorbic acid at 3.1 A resolution reveals the locations of three binding sites | | 分子名称: | ASCORBIC ACID, Hyaluronidase, phage associated | | 著者 | Mishra, P, Ethayathulla, A.S, Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Bhakuni, V, Singh, T.P. | | 登録日 | 2008-09-19 | | 公開日 | 2009-09-29 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Polysaccharide binding sites in hyaluronate lyase--crystal structures of native phage-encoded hyaluronate lyase and its complexes with ascorbic acid and lactose.
Febs J., 276, 2009
|
|
2NC2
 
 | |
3FRD
 
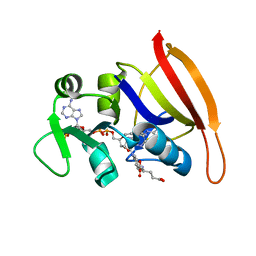 | | S. aureus DHFR complexed with NADPH and folate | | 分子名称: | DIHYDROFOLIC ACID, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Oefner, C, Dale-Glenn, E. | | 登録日 | 2009-01-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity
J.Antimicrob.Chemother., 63, 2009
|
|
3FRA
 
 | | Staphylococcus aureus F98Y DHFR complexed with iclaprim | | 分子名称: | 5-{[(2S)-2-cyclopropyl-7,8-dimethoxy-2H-chromen-5-yl]methyl}pyrimidine-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Oefner, C, Dale-Glenn, E. | | 登録日 | 2009-01-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity
J.Antimicrob.Chemother., 63, 2009
|
|
3FRE
 
 | | S. aureus DHFR complexed with NADPH and TMP | | 分子名称: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | 著者 | Oefner, C, Dale-Glenn, E. | | 登録日 | 2009-01-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity
J.Antimicrob.Chemother., 63, 2009
|
|
3FRB
 
 | | S. aureus F98Y DHFR complexed with TMP | | 分子名称: | Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIMETHOPRIM | | 著者 | Oefner, C, Dale-Glenn, E. | | 登録日 | 2009-01-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity
J.Antimicrob.Chemother., 63, 2009
|
|
3FRF
 
 | | S. aureus DHFR complexed with NADPH and iclaprim | | 分子名称: | 5-[[(2R)-2-cyclopropyl-7,8-dimethoxy-2H-chromen-5-yl]methyl]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Oefner, C, Dale-Glenn, E. | | 登録日 | 2009-01-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Increased hydrophobic interactions of iclaprim with Staphylococcus aureus dihydrofolate reductase are responsible for the increase in affinity and antibacterial activity
J.Antimicrob.Chemother., 63, 2009
|
|
2PYC
 
 | | Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution | | 分子名称: | ACETATE ION, ACETONITRILE, Phospholipase A2 VRV-PL-VIIIa, ... | | 著者 | Kumar, S, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | 登録日 | 2007-05-16 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of a monomeric phospholipase A2 from Russell's viper at 1.5A resolution
To be Published
|
|
2PT3
 
 | | Crystal structure of bovine lactoperoxidase at 2.34 A resolution reveals multiple anion binding sites | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | 著者 | Singh, A.K, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | 登録日 | 2007-05-08 | | 公開日 | 2007-05-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Crystal structure of bovine lactoperoxidase at 2.34 A resolution reveals multiple anion binding sites
To be Published
|
|
2W6Z
 
 | |
2W6O
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with 4-Amino-7,7-dimethyl-7,8-dihydro-quinazolinone fragment | | 分子名称: | 4-amino-7,7-dimethyl-7,8-dihydroquinazolin-5(6H)-one, BIOTIN CARBOXYLASE, CHLORIDE ION | | 著者 | Mochalkin, I, Miller, J.R. | | 登録日 | 2008-12-18 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W6Q
 
 | | Crystal structure of Biotin carboxylase from E. coli in complex with the triazine-2,4-diamine fragment | | 分子名称: | 6-(2-phenoxyethoxy)-1,3,5-triazine-2,4-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | 著者 | Mochalkin, I, Miller, J.R. | | 登録日 | 2008-12-18 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Antibacterial Biotin Carboxylase Inhibitors by Virtual Screening and Fragment-Based Approaches.
Acs Chem.Biol., 4, 2009
|
|
2W71
 
 | |
2W70
 
 | |
1FV0
 
 | | FIRST STRUCTURAL EVIDENCE OF THE INHIBITION OF PHOSPHOLIPASE A2 BY ARISTOLOCHIC ACID: CRYSTAL STRUCTURE OF A COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND ARISTOLOCHIC ACID | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, 9-HYDROXY ARISTOLOCHIC ACID, ACETATE ION, ... | | 著者 | Chandra, V, Jasti, J, Kaur, P, Srinivasan, A, Betzel, C, Singh, T.P. | | 登録日 | 2000-09-18 | | 公開日 | 2002-08-28 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Basis of Phospholipase A2 Inhibition for the Synthesis of Prostaglandins by the Plant Alkaloid Aristolochic Acid from a 1.7 A Crystal Structure
Biochemistry, 41, 2002
|
|
2W6P
 
 | |
2W6M
 
 | |
2W6N
 
 | |
3ZYU
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with I-BET151(GSK1210151A) | | 分子名称: | 1,2-ETHANEDIOL, 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, BROMODOMAIN-CONTAINING PROTEIN 4 | | 著者 | Chung, C.W, Mirguet, O. | | 登録日 | 2011-08-25 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Inhibition of Bet Recruitment to Chromatin as an Effective Treatment for Mll-Fusion Leukaemia.
Nature, 478, 2011
|
|
2CNB
 
 | | Trypanosoma brucei UDP-galactose-4-epimerase in ternary complex with NAD and the substrate analogue UDP-4-deoxy-4-fluoro-alpha-D-galactose | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GALACTOSE-4-EPIMERASE, URIDINE-5'-DIPHOSPHATE-4-DEOXY-4-FLUORO-ALPHA-D-GALACTOSE | | 著者 | Alphey, M.S, Ferguson, M.A.J, Hunter, W.N. | | 登録日 | 2006-05-18 | | 公開日 | 2006-06-12 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Trypanosoma Brucei Udp-Galactose-4-Epimerase in Ternary Complex with Nad+ and the Substrate Analogue Udp-4-Deoxy-4-Fluoro-Alpha-D-Galactose
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3MEZ
 
 | | X-ray structural analysis of a mannose specific lectin from dutch crocus (crocus vernus) | | 分子名称: | FORMIC ACID, GLYCEROL, Mannose-specific lectin 3 chain 1, ... | | 著者 | Akrem, A, Meyer, A, Perbandt, M, Voelter, W, Buck, F, Betzel, C. | | 登録日 | 2010-04-01 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | X-ray structural analysis of a mannose specific lectin from dutch crocus (crocus vernus)
To be Published
|
|
3IFM
 
 | |
5EYU
 
 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-25 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
