5XM8
 
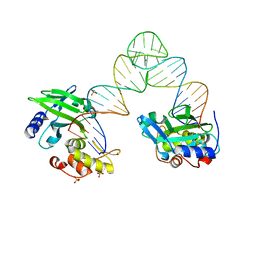 | |
5XMA
 
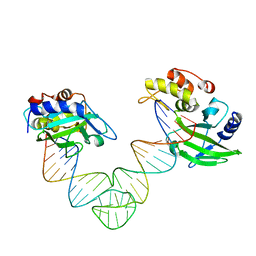 | |
5XM9
 
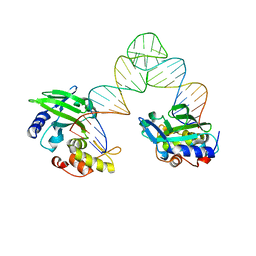 | |
5GL6
 
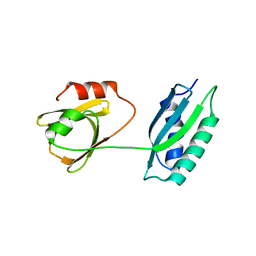 | | Msmeg rimP | | 分子名称: | Ribosome maturation factor RimP | | 著者 | Chu, T, Lau, T.C.K. | | 登録日 | 2016-07-08 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The ribosomal maturation factor P fromMycobacterium smegmatisfacilitates the ribosomal biogenesis by binding to the small ribosomal protein S12.
J. Biol. Chem., 294, 2019
|
|
5GO9
 
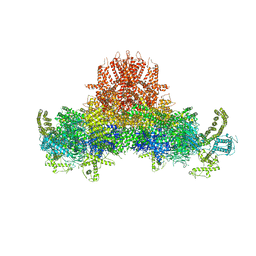 | |
5GS9
 
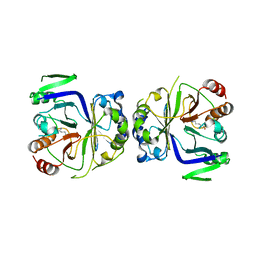 | | Crystal structure of CASTOR1-arginine | | 分子名称: | ARGININE, GATS-like protein 3 | | 著者 | Zhang, T, Ding, J. | | 登録日 | 2016-08-15 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling.
Cell Discov, 2, 2016
|
|
5TZ2
 
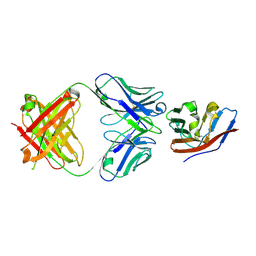 | | Crystal structure of human CD47 ECD bound to Fab of C47B222 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C47B222 Fab Heavy Chain, ... | | 著者 | Cardoso, R.M.F. | | 登録日 | 2016-11-21 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
5TZT
 
 | |
5TZU
 
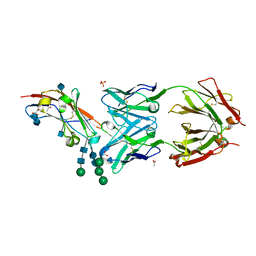 | | Crystal structure of human CD47 ECD bound to Fab of B6H12.2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Cardoso, R.M.F. | | 登録日 | 2016-11-22 | | 公開日 | 2017-03-15 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
5YR0
 
 | | Structure of Beclin1-UVRAG coiled coil domain complex | | 分子名称: | Beclin-1, UV radiation resistance associated protein | | 著者 | Pan, X, Zhao, Y, He, Y. | | 登録日 | 2017-11-08 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5GOA
 
 | |
7KL9
 
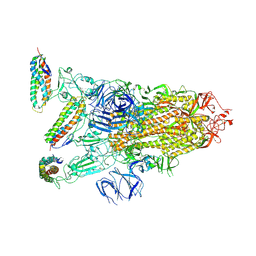 | |
7KJS
 
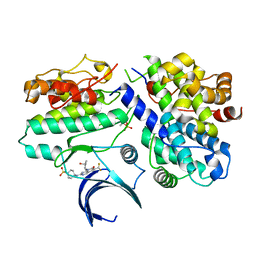 | | Crystal structure of CDK2/cyclin E in complex with PF-06873600 | | 分子名称: | 6-(difluoromethyl)-8-[(1R,2R)-2-hydroxy-2-methylcyclopentyl]-2-{[1-(methylsulfonyl)piperidin-4-yl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | 著者 | McTigue, M.A, He, Y, Ferre, R.A. | | 登録日 | 2020-10-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Discovery of PF-06873600, a CDK2/4/6 Inhibitor for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
4WIP
 
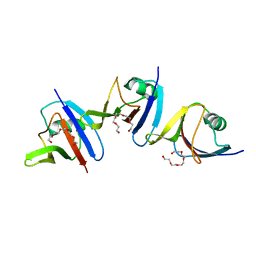 | | DIX domain of human Dvl2 | | 分子名称: | PENTAETHYLENE GLYCOL, Segment polarity protein dishevelled homolog DVL-2 | | 著者 | Fiedler, M, Bienz, M, Madrzak, J, Chin, J.W. | | 登録日 | 2014-09-26 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.691 Å) | | 主引用文献 | Ubiquitination of the Dishevelled DIX domain blocks its head-to-tail polymerization.
Nat Commun, 6, 2015
|
|
6A85
 
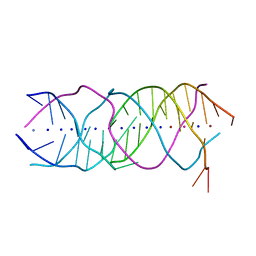 | | Crystal structure of a novel DNA quadruplex | | 分子名称: | AMMONIUM ION, DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*TP*GP*GP*GP*TP*GP*CP*GP*TP*T)-3'), LEAD (II) ION, ... | | 著者 | Liu, H.H, Gan, J.H. | | 登録日 | 2018-07-06 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | High-resolution DNA quadruplex structure containing all the A-, G-, C-, T-tetrads.
Nucleic Acids Res., 46, 2018
|
|
7XS8
 
 | |
7XSC
 
 | |
4X3V
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | 分子名称: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | 著者 | Dealwis, C.G, Ahmad, M.F, Alam, I. | | 登録日 | 2014-12-01 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
7XSA
 
 | |
7XSB
 
 | |
6BJX
 
 | |
3MEE
 
 | | HIV-1 Reverse Transcriptase in Complex with TMC278 | | 分子名称: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
3MEG
 
 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC278 | | 分子名称: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
3MED
 
 | | HIV-1 K103N Reverse Transcriptase in Complex with TMC125 | | 分子名称: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, CHLORIDE ION, SULFATE ION, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
3MEC
 
 | | HIV-1 Reverse Transcriptase in Complex with TMC125 | | 分子名称: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, SULFATE ION, p51 Reverse transcriptase, ... | | 著者 | Lansdon, E.B. | | 登録日 | 2010-03-31 | | 公開日 | 2010-05-12 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
